You are browsing environment: FUNGIDB
CAZyme Information: H257_04760-t26_3-p1
You are here: Home > Sequence: H257_04760-t26_3-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aphanomyces astaci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Saprolegniaceae; Aphanomyces; Aphanomyces astaci | |||||||||||
| CAZyme ID | H257_04760-t26_3-p1 | |||||||||||
| CAZy Family | GH17|CBM13|CBM13 | |||||||||||
| CAZyme Description | hypothetical protein, variant 3 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 953233; End:955153 Strand: - | |||||||||||
Full Sequence Download help
| MKGSPSATLS PLWQGTVPLL CVALLAFAVF ANTLSCGFVW DDRAAILTNR DIRTDDNTTA | 60 |
| VGDLFVHDFW GTPISSASSH KSFRPITVLT FRLNYAVGGF DPWGYHLLNV VLHSITSALV | 120 |
| VVVGRRVMSI APSSSQVHRA PVLAGLLFAV HPIHCDSVAS VVGRADVLCT LVSLVAFTCY | 180 |
| DTAISDRTTT KWMHFILSIG LIVLATLCKE LGATTLGMLV VLDLLQSYRL STQELLASPT | 240 |
| LGRLAVLVAF GTTAIAGRIL LNGPHMLYPW TEMENDISLL PFGTSKVLTI AHTHAWYLYK | 300 |
| MAWPQYLSYD YGFRTIPIVH SMLDPRNIST AIAYTCVVTL VFASAWHSRT SPSLLLMASF | 360 |
| ALFPFVPAAN VLFPVGTIVA ERLLYFPSVG VCLLAGYTLD TALHRGSQRQ QVALVGIATT | 420 |
| LLIVATARTL CRNRDWTDEV ALFEASVKVA PWSTKVLSNL SKVLLNSDAP RAAAYLERAL | 480 |
| HVLPHYSIGH FNLGLAYVNM GKSLHCMDSL LKAIDVDHSL ASYSYLGKYL FEFYATNQHD | 540 |
| KFRTDQPTHA LATATKLLDF TLSHHHNLPT IVFTRGLIAY YADDFAAALP YVVAYRQLCL | 600 |
| QH | 602 |
Enzyme Prediction help
| EC | 2.4.1.109:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT105 | 93 | 233 | 3e-36 | 0.9694656488549618 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400627 | DUF1736 | 9.45e-11 | 270 | 338 | 6 | 74 | Domain of unknown function (DUF1736). This domain of unknown function is found in various hypothetical metazoan proteins. |
| 276809 | TPR | 3.89e-08 | 427 | 515 | 8 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 1.52e-06 | 455 | 532 | 2 | 81 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 274350 | PEP_TPR_lipo | 1.63e-05 | 404 | 544 | 746 | 892 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
| 274350 | PEP_TPR_lipo | 0.004 | 426 | 566 | 335 | 470 | putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAD7262601.1|GT105 | 5.20e-78 | 24 | 513 | 13 | 506 |
| BAE38405.1|GT105 | 3.71e-73 | 28 | 503 | 23 | 501 |
| BAC32122.1|GT105 | 8.35e-73 | 28 | 503 | 23 | 501 |
| AAI66274.1|GT105 | 1.71e-71 | 5 | 486 | 12 | 511 |
| CAG26973.1|GT105|2.4.1.109 | 3.14e-70 | 28 | 513 | 18 | 506 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q8BRH0|TMTC3_MOUSE | 1.75e-72 | 28 | 503 | 23 | 501 | Protein O-mannosyl-transferase TMTC3 OS=Mus musculus OX=10090 GN=Tmtc3 PE=1 SV=2 |
| sp|Q6ZXV5|TMTC3_HUMAN | 5.65e-71 | 28 | 513 | 18 | 506 | Protein O-mannosyl-transferase TMTC3 OS=Homo sapiens OX=9606 GN=TMTC3 PE=1 SV=2 |
| sp|Q5T4D3|TMTC4_HUMAN | 1.18e-70 | 5 | 473 | 13 | 501 | Protein O-mannosyl-transferase TMTC4 OS=Homo sapiens OX=9606 GN=TMTC4 PE=1 SV=2 |
| sp|Q7K4B6|TMTC3_DROME | 4.11e-69 | 7 | 502 | 26 | 563 | Protein O-mannosyl-transferase Tmtc3 OS=Drosophila melanogaster OX=7227 GN=Tmtc3 PE=2 SV=1 |
| sp|Q8BG19|TMTC4_MOUSE | 5.99e-69 | 7 | 473 | 14 | 501 | Protein O-mannosyl-transferase TMTC4 OS=Mus musculus OX=10090 GN=Tmtc4 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
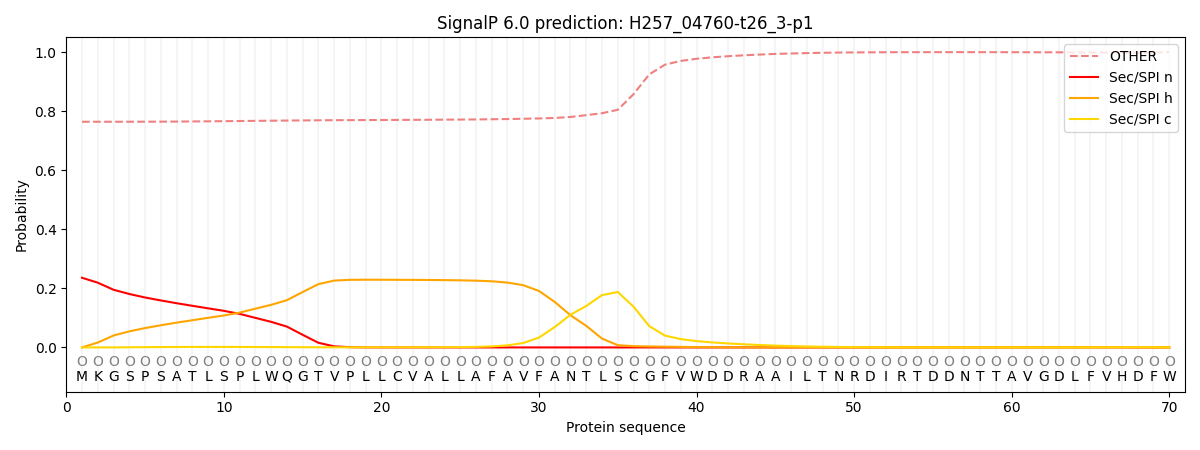
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.773434 | 0.226567 |
TMHMM Annotations download full data without filtering help
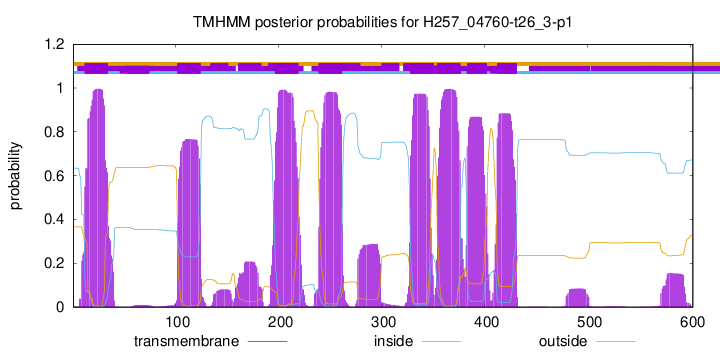
| Start | End |
|---|---|
| 12 | 34 |
| 102 | 124 |
| 197 | 219 |
| 239 | 261 |
| 328 | 350 |
| 354 | 376 |
| 383 | 402 |
| 412 | 431 |
