You are browsing environment: FUNGIDB
CAZyme Information: GLOIN_2v1705027-t44_1-p1
You are here: Home > Sequence: GLOIN_2v1705027-t44_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | GLOIN_2v1705027-t44_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | glycosyltransferase family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 260 | 462 | 5.9e-29 | 0.4397905759162304 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 3.77e-47 | 43 | 483 | 2 | 404 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 1.12e-29 | 54 | 539 | 11 | 483 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 223071 | egt | 2.31e-25 | 230 | 461 | 238 | 441 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 224732 | YjiC | 7.26e-17 | 44 | 481 | 4 | 396 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 177813 | PLN02152 | 6.45e-09 | 245 | 465 | 198 | 432 | indole-3-acetate beta-glucosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.01e-54 | 3 | 553 | 2 | 544 | |
| 6.36e-54 | 42 | 519 | 9 | 502 | |
| 8.83e-53 | 42 | 518 | 10 | 502 | |
| 1.66e-52 | 42 | 534 | 9 | 515 | |
| 1.66e-52 | 42 | 534 | 9 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.91e-14 | 272 | 462 | 19 | 186 | Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_B Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_C Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens],6IPB_D Crystal Structure of the Cofactor-binding Domain of the Human Phase II Drug Metabolism Enzyme UGT2B15 [Homo sapiens] |
|
| 6.81e-13 | 275 | 461 | 1 | 164 | Chain A, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_B Chain B, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_C Chain C, UDP-glucuronosyltransferase 2B15 [Homo sapiens],7CJX_D Chain D, UDP-glucuronosyltransferase 2B15 [Homo sapiens] |
|
| 1.86e-12 | 281 | 462 | 6 | 164 | Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens],2O6L_B Crystal Structure of the UDP-Glucuronic Acid Binding Domain of the Human Drug Metabolizing UDP-Glucuronosyltransferase 2B7 [Homo sapiens] |
|
| 7.92e-09 | 367 | 481 | 284 | 395 | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering [Streptomyces antibioticus],2IYF_B The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering [Streptomyces antibioticus] |
|
| 7.92e-09 | 367 | 481 | 284 | 395 | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A [Streptomyces antibioticus],4M83_B Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A [Streptomyces antibioticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.78e-22 | 54 | 530 | 34 | 505 | UDP-glucuronosyltransferase 2B23 OS=Macaca fascicularis OX=9541 GN=UGT2B23 PE=1 SV=1 |
|
| 3.75e-22 | 137 | 530 | 132 | 501 | UDP-glucuronosyltransferase 1A7 OS=Mus musculus OX=10090 GN=Ugt1a7 PE=1 SV=1 |
|
| 4.96e-22 | 54 | 530 | 34 | 505 | UDP-glucuronosyltransferase 2B9 OS=Macaca fascicularis OX=9541 GN=UGT2B9 PE=2 SV=1 |
|
| 8.84e-22 | 239 | 530 | 245 | 505 | UDP-glucuronosyltransferase 2B33 OS=Macaca mulatta OX=9544 GN=UGT2B33 PE=1 SV=1 |
|
| 1.57e-20 | 239 | 530 | 245 | 505 | UDP-glucuronosyltransferase 2B4 OS=Homo sapiens OX=9606 GN=UGT2B4 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
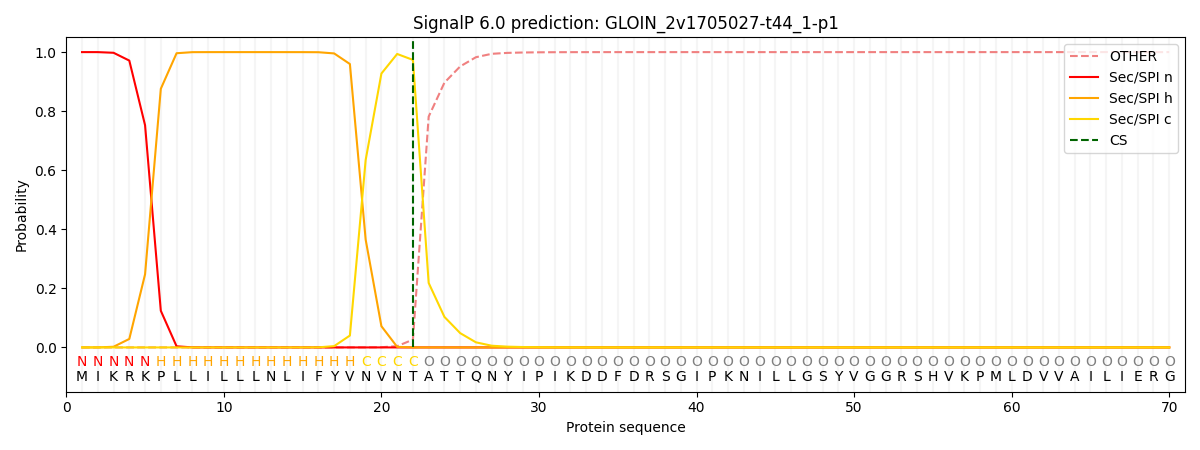
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000238 | 0.999726 | CS pos: 22-23. Pr: 0.9736 |

