You are browsing environment: FUNGIDB
CAZyme Information: GAQ11698.1
You are here: Home > Sequence: GAQ11698.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ11698.1 | |||||||||||
| CAZy Family | PL4 | |||||||||||
| CAZyme Description | putative HC-toxin efflux carrier TOXA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 269892 | GH36 | 6.87e-43 | 260 | 466 | 5 | 210 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 341045 | MFS_Azr1_MDR_like | 1.03e-30 | 578 | 729 | 167 | 319 | Saccharomyces cerevisiae Azole resistance protein 1 (Azr1p), and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This subfamily is composed of multidrug resistance (MDR) transporters including various Saccharomyces cerevisiae proteins such as azole resistance protein 1 (Azr1p), vacuolar basic amino acid transporter 1 (Vba1p), vacuolar basic amino acid transporter 5 (Vba5p), and Sge1p (also known as Nor1p, 10-N-nonyl acridine orange resistance protein, and crystal violet resistance protein). MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. This subfamily belongs to the Methylenomycin A resistance protein (also called MMR peptide) and similar multidrug resistance (MDR) transporters (MMR-like MDR transporter) family of the Major Facilitator Superfamily (MFS) of transporters. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 307952 | Melibiase | 2.25e-15 | 218 | 465 | 1 | 250 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| 340880 | MFS_ARN_like | 5.71e-12 | 509 | 733 | 215 | 443 | Yeast ARN family of Siderophore iron transporters and similar proteins of the Major Facilitator Superfamily. The ARN family of siderophore iron transporters includes ARN1 (or ferrichrome permease), ARN2 (or triacetylfusarinine C transporter 1 or TAF1), ARN3 (or siderophore iron transporter 1 or SIT1 or ferrioxamine B permease) and ARN4 (or Enterobactin permease or ENB1). They specifically recognize siderophore-iron chelates are expressed under conditions of iron deprivation. They facilitate the uptake of both hydroxamate- and catecholate-type siderophores. This group also includes glutathione exchanger 1 (Gex1p) and Gex2p, which are proton/glutathione antiporters that import glutathione from the vacuole and exports it through the plasma membrane. The ARN family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 225882 | GalA | 7.58e-12 | 271 | 472 | 305 | 504 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.48e-129 | 36 | 494 | 33 | 548 | |
| 1.29e-128 | 1 | 477 | 1 | 538 | |
| 6.07e-128 | 1 | 477 | 58 | 595 | |
| 6.07e-128 | 1 | 477 | 58 | 595 | |
| 3.97e-127 | 1 | 477 | 1 | 538 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.15e-10 | 244 | 461 | 320 | 538 | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN0_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN1_A Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_C Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_D Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM] |
|
| 8.15e-10 | 244 | 461 | 320 | 538 | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose [Lactobacillus acidophilus NCFM] |
|
| 8.22e-10 | 94 | 461 | 131 | 535 | Chain A, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_B Chain B, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_C Chain C, Alpha-galactosidase [Levilactobacillus brevis ATCC 367],3MI6_D Chain D, Alpha-galactosidase [Levilactobacillus brevis ATCC 367] |
|
| 1.67e-08 | 89 | 465 | 139 | 551 | SpAga wild type apo structure [Streptococcus pneumoniae TIGR4],6PHV_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4] |
|
| 6.56e-08 | 89 | 465 | 139 | 551 | Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PHX_A SpAga D472N structure in complex with raffinose [Streptococcus pneumoniae TIGR4],6PHY_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PI0_A AgaD472N-Linear Blood group B type 2 trisaccharide complex structure [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.41e-72 | 465 | 806 | 165 | 529 | MFS-type efflux pump MFS1 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=MFS1 PE=3 SV=2 |
|
| 4.41e-72 | 465 | 806 | 165 | 529 | MFS-type efflux pump MFS1 OS=Trichophyton rubrum (strain ATCC MYA-4607 / CBS 118892) OX=559305 GN=MFS1 PE=2 SV=2 |
|
| 1.61e-67 | 503 | 804 | 223 | 526 | Efflux pump roqT OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=roqT PE=1 SV=1 |
|
| 2.70e-67 | 464 | 815 | 119 | 494 | MFS-type transporter nsrN OS=Aspergillus novofumigatus (strain IBT 16806) OX=1392255 GN=nsrN PE=3 SV=1 |
|
| 2.80e-67 | 464 | 806 | 208 | 572 | MFS-type transporter cdmB OS=Talaromyces verruculosus OX=198730 GN=cdmB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000000 |
TMHMM Annotations download full data without filtering help
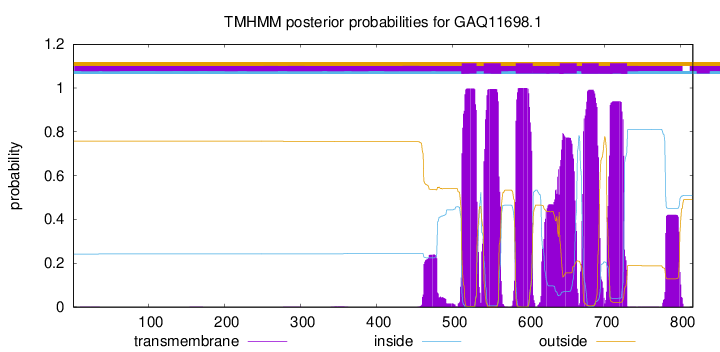
| Start | End |
|---|---|
| 512 | 531 |
| 541 | 563 |
| 583 | 605 |
| 641 | 663 |
| 670 | 692 |
| 707 | 729 |
