You are browsing environment: FUNGIDB
CAZyme Information: GAQ11411.1
You are here: Home > Sequence: GAQ11411.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ11411.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | probable glucan 1,3-beta-glucosidase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:29 | 3.2.1.21:6 | 3.2.1.75:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 90 | 386 | 1.1e-110 | 0.9966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 2.21e-67 | 41 | 415 | 17 | 394 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 9.80e-17 | 92 | 311 | 19 | 238 | Cellulase (glycosyl hydrolase family 5). |
| 277321 | MPP_ASMase | 0.003 | 88 | 206 | 141 | 251 | acid sphingomyelinase and related proteins, metallophosphatase domain. Acid sphingomyelinase (ASMase) is a ubiquitously expressed phosphodiesterase which hydrolyzes sphingomyelin in acid pH conditions to form ceramide, a bioactive second messenger, as part of the sphingomyelin signaling pathway. ASMase is localized at the noncytosolic leaflet of biomembranes (for example the luminal leaflet of endosomes, lysosomes and phagosomes, and the extracellular leaflet of plasma membranes). ASMase-deficient humans develop Niemann-Pick disease. This disease is characterized by lysosomal storage of sphingomyelin in all tissues. Although ASMase-deficient mice are resistant to stress-induced apoptosis, they have greater susceptibility to bacterial infection. The latter correlates with defective phagolysosomal fusion and antibacterial killing activity in ASMase-deficient macrophages. ASMase belongs to the metallophosphatase (MPP) superfamily. MPPs are functionally diverse, but all share a conserved domain with an active site consisting of two metal ions (usually manganese, iron, or zinc) coordinated with octahedral geometry by a cage of histidine, aspartate, and asparagine residues. The MPP superfamily includes: the phosphoprotein phosphatases (PPPs), Mre11/SbcD-like exonucleases, Dbr1-like RNA lariat debranching enzymes, YfcE-like phosphodiesterases, purple acid phosphatases (PAPs), YbbF-like UDP-2,3-diacylglucosamine hydrolases, and acid sphingomyelinases (ASMases). The conserved domain is a double beta-sheet sandwich with a di-metal active site made up of residues located at the C-terminal side of the sheets. This domain is thought to allow for productive metal coordination. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.83e-298 | 1 | 416 | 1 | 416 | |
| 1.14e-200 | 7 | 416 | 7 | 416 | |
| 1.14e-200 | 7 | 416 | 7 | 416 | |
| 2.93e-196 | 1 | 416 | 1 | 416 | |
| 2.53e-192 | 1 | 415 | 1 | 415 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.54e-122 | 36 | 415 | 2 | 391 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 9.11e-122 | 36 | 415 | 2 | 391 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 1.11e-121 | 36 | 415 | 8 | 397 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 3.15e-121 | 36 | 415 | 8 | 397 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
|
| 3.15e-121 | 36 | 415 | 8 | 397 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.60e-313 | 1 | 416 | 1 | 416 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgA PE=3 SV=1 |
|
| 4.31e-300 | 1 | 416 | 1 | 416 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
|
| 5.03e-299 | 1 | 416 | 1 | 416 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
|
| 1.30e-248 | 1 | 416 | 1 | 415 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=exgA PE=3 SV=2 |
|
| 5.20e-197 | 1 | 416 | 1 | 416 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
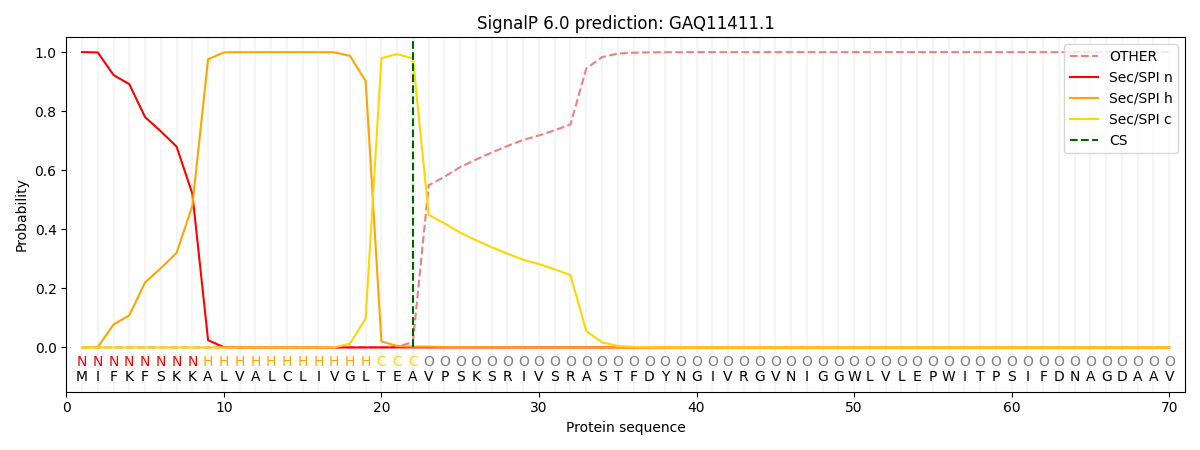
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000299 | 0.999676 | CS pos: 22-23. Pr: 0.9786 |
