You are browsing environment: FUNGIDB
CAZyme Information: GAQ06282.1
You are here: Home > Sequence: GAQ06282.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
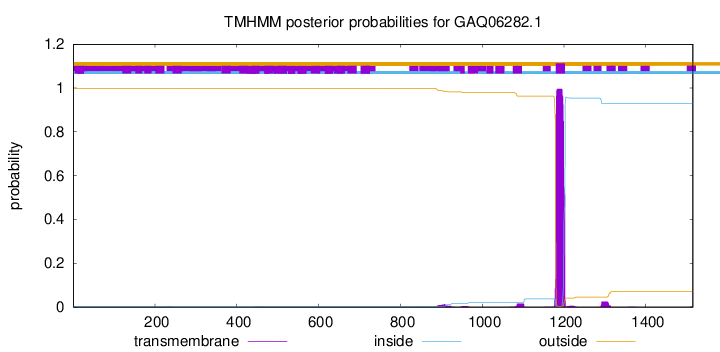
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ06282.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | serine/threonine-protein kinase cds1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1033083; End:1039127 Strand: + | |||||||||||
Full Sequence Download help
| MEATQESTQP CTDPRRIGRN NSGLLEEDVS DIICILHPTS LAAHEAVAAT ASLAPQHILQ | 60 |
| KDELEYEDPD TAALDIALRL SSNVRNINLG FCFGRNRSRC DLLLAPDDNA KRISNTHFRI | 120 |
| HLTGDGILML EDLSTNGTVV DDCRLRKNQK ENSRMLTNGS VIQVLRGNNA SDEVRFVVRI | 180 |
| PSRDGYAMRY TENMLRYLER VQRQPAGAMQ KNRQGSVRPS LEWTVANAYG MHWTGGSMYN | 240 |
| VTGQIGKGAF ATVYKLATKQ HGAVYAAKEL DKRRFMKNGI LDQKVDNEMK IMKDLKHPNI | 300 |
| VQYIDHHEHD RWIYIIMEYV PGGELSTYLS THGKIPEDMV KMLARQLLHA LQYLHKRRIT | 360 |
| HRDIKPDNIL IASLEPLRVK LSDFGLSKVV QEETFLKTFC GTLLYCAPEV YPEYENYRRG | 420 |
| EVRKRRRLGD PPPKTSPYDQ SVDMWSFGAV LFHILSGTPP YTGRGDDRGS QMLRNIMTTD | 480 |
| PDYDVLRREG VSEAGVDFVR RLLNRDPHSR PKESECFQHS WIRDVPDVDE YDDNDIQPAD | 540 |
| FGALSDIGED LENELDASQI SLNDNPEPVL TGDESHDSND LAQSKRPRID HLPADIHYPS | 600 |
| LPNIESFPAA RPIPETTPRR LFGEITSSAL RSSNVFGASM NAFGGDDLSV HDFVSSTGES | 660 |
| IISDGNSLNS VLSLPDNPFA GSAPSLMGAE NLVGQLNMNS WHPGTSAHGP PAATELPALK | 720 |
| TSVDEGVGGS PGPELNKETH PSTSTPKGAK FSRRIELPLP DTASERSSRE ITRENSKAPS | 780 |
| DKPEASAGEV FDIELAVTID ARTGRQISHP PEGIVSDTTS AALRHEMPTE IPPTISLHRH | 840 |
| EQPRPLLGKL TTLPGSIFDL TIRLEDRMTS WGRGPLATIC HPDPMDTRIP AYALEVTFWA | 900 |
| PAIEAQIAAG RNWMEVPGVM AILSTKTRKC IWVNDTELRR GSEGSSTREG FHFGKLYTGD | 960 |
| IITIYRHRNK FLKFQCEFYH GDSARSRPEE EKGFVVRKVL MSKEGVAANR LPVRKDNDGG | 1020 |
| KKQGINPKTS AIYENLRADL FTIFNLPDGL DTPMSLSRSP SPHPGGGWSS PGLTPGSGTS | 1080 |
| TPRSGFLSPN SLGPSGISWA AARAKSDEVR GYPSFSTRNN GFFSRSKRKI TSSLPRFRMN | 1140 |
| GSARNGYVDK DEYWRGHNSS EAGLRFGFAR GLMRRRRSRA FLALIVLVLG YLFFWTAIVQ | 1200 |
| SYRRSSFGGG RKFVIILPSN VEGGVMEWKG AREWAIERNS ISNKEEYAKR WGYELEIVNM | 1260 |
| LAKKRYSHEW RESWEKVDII RDAMRKYPNA EWFWWLDLNT WIMEYSYSLQ DHIFDRLGEL | 1320 |
| TYRDINLYNP LNISHPPTAP YLDELSRSAE GDGNPSSIEL LLSQDCGGFN LGSFFIKRSL | 1380 |
| WSDRLMDLWW DPVMYEQKHM DWEHKEQDAL EYLYQSQPWI RSNVAFTPQR YINSFPPGAC | 1440 |
| GEGGDPDVHY SVKDRDFMVN MAGCQFGRDC WGEMYLYREL SKKLNRTRWQ RLKDGLGELY | 1500 |
| SRLLPKEDKP QEQQQ | 1515 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 1212 | 1472 | 1.7e-85 | 0.983739837398374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214567 | S_TKc | 4.33e-86 | 239 | 522 | 1 | 254 | Serine/Threonine protein kinases, catalytic domain. Phosphotransferases. Serine or threonine-specific kinase subfamily. |
| 270687 | STKc_CAMK | 2.30e-83 | 239 | 521 | 2 | 258 | The catalytic domain of CAMK family Serine/Threonine Kinases. STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. CaMKs are multifunctional calcium and calmodulin (CaM) stimulated STKs involved in cell cycle regulation. There are several types of CaMKs including CaMKI, CaMKII, and CaMKIV. CaMKI proteins are monomeric and they play pivotal roles in the nervous system, including long-term potentiation, dendritic arborization, neurite outgrowth, and the formation of spines, synapses, and axons. CaMKII is a signaling molecule that translates upstream calcium and reactive oxygen species (ROS) signals into downstream responses that play important roles in synaptic function and cardiovascular physiology. CAMKIV is implicated in regulating several transcription factors like CREB, MEF2, and retinoid orphan receptors, as well as in T-cell development and signaling. The CAMK family also consists of other related kinases including the Phosphorylase kinase Gamma subunit (PhKG), the C-terminal kinase domains of Ribosomal S6 kinase (RSK) and Mitogen and stress-activated kinase (MSK), Doublecortin-like kinase (DCKL), and the MAPK-activated protein kinases MK2, MK3, and MK5, among others. The CAMK family is part of a larger superfamily that includes the catalytic domains of other protein STKs, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. |
| 368535 | Glyco_transf_34 | 2.68e-80 | 1212 | 1470 | 4 | 238 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 271000 | STKc_Rad53_Cds1 | 3.11e-74 | 239 | 521 | 2 | 265 | Catalytic domain of the yeast Serine/Threonine Kinases, Rad53 and Cds1. STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. Rad53 and Cds1 are the checkpoint kinase 2 (Chk2) homologs found in budding and fission yeast, respectively. They play a central role in the cell's response to DNA lesions to prevent genome rearrangements and maintain genome integrity. They are phosphorylated in response to DNA damage and incomplete replication, and are essential for checkpoint control. They help promote DNA repair by stalling the cell cycle prior to mitosis in the presence of DNA damage. The Rad53/Cds1 subfamily is part of a larger superfamily that includes the catalytic domains of other protein STKs, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. |
| 270783 | STKc_MAPKKK | 5.75e-70 | 244 | 522 | 7 | 258 | Catalytic domain of the Serine/Threonine Kinase, Mitogen-Activated Protein Kinase Kinase Kinase. STKs catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. MAPKKKs (MKKKs or MAP3Ks) are also called MAP/ERK kinase kinases (MEKKs) in some cases. They phosphorylate and activate MAPK kinases (MAPKKs or MKKs or MAP2Ks), which in turn phosphorylate and activate MAPKs during signaling cascades that are important in mediating cellular responses to extracellular signals. This subfamily is composed of the Apoptosis Signal-regulating Kinases ASK1 (or MAPKKK5) and ASK2 (or MAPKKK6), MEKK1, MEKK2, MEKK3, MEKK4, as well as plant and fungal MAPKKKs. Also included in this subfamily are the cell division control proteins Schizosaccharomyces pombe Cdc7 and Saccharomyces cerevisiae Cdc15. The MAPKKK subfamily is part of a larger superfamily that includes the catalytic domains of other STKs, protein tyrosine kinases, RIO kinases, aminoglycoside phosphotransferase, choline kinase, and phosphoinositide 3-kinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRD89754.1|GT34 | 9.26e-233 | 1054 | 1508 | 1 | 458 |
| QMW42907.1|GT34 | 9.26e-233 | 1054 | 1508 | 1 | 458 |
| QMW30853.1|GT34 | 9.26e-233 | 1054 | 1508 | 1 | 458 |
| UDD59694.1|GT34 | 9.26e-233 | 1054 | 1508 | 1 | 458 |
| CAK42408.1|GT34 | 1.88e-231 | 1054 | 1508 | 1 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XZV_A | 1.81e-62 | 85 | 524 | 60 | 468 | Crystal structure of Rad53 1-466 in complex with AMP-PNP [Saccharomyces cerevisiae S288C],5XZV_B Crystal structure of Rad53 1-466 in complex with AMP-PNP [Saccharomyces cerevisiae S288C],5XZW_A Crystal structure of Rad53 1-466 [Saccharomyces cerevisiae S288C],5XZW_B Crystal structure of Rad53 1-466 [Saccharomyces cerevisiae S288C] |
| 4PDP_A | 5.27e-51 | 245 | 526 | 39 | 305 | Crystal structure of Rad53 kinase domain and SCD2 [Saccharomyces cerevisiae],4PDP_B Crystal structure of Rad53 kinase domain and SCD2 [Saccharomyces cerevisiae],4PDS_A Crystal structure of Rad53 kinase domain and SCD2 in complex with AMPPNP [Saccharomyces cerevisiae],4PDS_B Crystal structure of Rad53 kinase domain and SCD2 in complex with AMPPNP [Saccharomyces cerevisiae] |
| 3BHY_A | 2.63e-44 | 239 | 525 | 7 | 272 | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with a beta-carboline ligand [Homo sapiens],3BQR_A Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with an imidazo-pyridazine ligand [Homo sapiens],5A6N_A Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 [Homo sapiens],5A6N_B Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 [Homo sapiens],5A6O_A Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) [Homo sapiens],5A6O_B Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) [Homo sapiens],5VJA_A Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) [Homo sapiens],5VJA_B Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) [Homo sapiens],5VJA_C Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) [Homo sapiens],5VJA_D Crystal Structure of human zipper-interacting protein kinase (ZIPK, alias DAPK3) in complex with a pyrazolo[3,4-d]pyrimidinone ligand (HS38) [Homo sapiens] |
| 1YRP_A | 3.11e-44 | 239 | 523 | 14 | 277 | Catalytic domain of human ZIP kinase phosphorylated at Thr265 [Homo sapiens],1YRP_B Catalytic domain of human ZIP kinase phosphorylated at Thr265 [Homo sapiens] |
| 2J90_A | 4.68e-44 | 239 | 525 | 28 | 293 | Crystal structure of human ZIP kinase in complex with a tetracyclic pyridone inhibitor (Pyridone 6) [Homo sapiens],2J90_B Crystal structure of human ZIP kinase in complex with a tetracyclic pyridone inhibitor (Pyridone 6) [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P50108|MNN10_YEAST | 2.35e-106 | 1203 | 1491 | 112 | 389 | Probable alpha-1,6-mannosyltransferase MNN10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN10 PE=1 SV=1 |
| sp|P78817|YFE6_SCHPO | 3.29e-85 | 1214 | 1492 | 76 | 343 | Uncharacterized alpha-1,2-galactosyltransferase C637.06 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC637.06 PE=2 SV=2 |
| sp|P22216|RAD53_YEAST | 3.70e-59 | 85 | 526 | 60 | 470 | Serine/threonine-protein kinase RAD53 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=RAD53 PE=1 SV=1 |
| sp|Q09170|CDS1_SCHPO | 2.68e-53 | 45 | 525 | 6 | 436 | Serine/threonine-protein kinase cds1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=cds1 PE=1 SV=2 |
| sp|Q54MH0|FHKD_DICDI | 2.80e-44 | 239 | 524 | 199 | 474 | Probable serine/threonine-protein kinase fhkD OS=Dictyostelium discoideum OX=44689 GN=fhkD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000067 | 0.000000 |