You are browsing environment: FUNGIDB
CAZyme Information: GAQ05995.1
You are here: Home > Sequence: GAQ05995.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ05995.1 | |||||||||||
| CAZy Family | GH15|CBM20 | |||||||||||
| CAZyme Description | solute carrier family 35 member E3 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT25 | 62 | 285 | 2e-29 | 0.988950276243094 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 293786 | ANK | 5.42e-14 | 762 | 871 | 2 | 98 | ankyrin repeats. Ankyrin repeats are one of the most abundant repeat motifs, and generally function as scaffolds for protein-protein interactions in processes including cell cycle, transcriptional regulation, signal transduction, vesicular trafficking, and inflammatory response. Although predominantly found in eukaryotic proteins, they are also found in some bacterial and viral proteins. Less is known of their physiological roles in prokaryotes. Some bacterial ANK proteins play key roles in microbial pathogenesis by mimicking or manipulating host function(s). The pathogen Providencia alcalifaciens N-formyltransferase ankyrin repeats function in small molecule binding and allosteric control. Ankyrin-repeat proteins have been associated with a number of human diseases. |
| 133474 | Glyco_transf_25 | 8.52e-14 | 64 | 152 | 1 | 96 | Glycosyltransferase family 25 [lipooligosaccharide (LOS) biosynthesis protein] is a family of glycosyltransferases involved in LOS biosynthesis. The members include the beta(1,4) galactosyltransferases: Lgt2 of Moraxella catarrhalis, LgtB and LgtE of Neisseria gonorrhoeae and Lic2A of Haemophilus influenzae. M. catarrhalis Lgt2 catalyzes the addition of galactose (Gal) to the growing chain of LOS on the cell surface. N. gonorrhoeae LgtB and LgtE link Gal-beta(1,4) to GlcNAc (N-acetylglucosamine) and Glc (glucose), respectively. The genes encoding LgtB and LgtE are two genes of a five gene locus involved in the synthesis of gonococcal LOS. LgtE is believed to perform the first step in LOS biosynthesis. |
| 223769 | RhaT | 4.29e-12 | 431 | 722 | 5 | 289 | Permease of the drug/metabolite transporter (DMT) superfamily [Carbohydrate transport and metabolism, Amino acid transport and metabolism, General function prediction only]. |
| 403870 | Ank_2 | 2.26e-11 | 769 | 871 | 3 | 91 | Ankyrin repeats (3 copies). |
| 293786 | ANK | 2.43e-09 | 797 | 874 | 3 | 68 | ankyrin repeats. Ankyrin repeats are one of the most abundant repeat motifs, and generally function as scaffolds for protein-protein interactions in processes including cell cycle, transcriptional regulation, signal transduction, vesicular trafficking, and inflammatory response. Although predominantly found in eukaryotic proteins, they are also found in some bacterial and viral proteins. Less is known of their physiological roles in prokaryotes. Some bacterial ANK proteins play key roles in microbial pathogenesis by mimicking or manipulating host function(s). The pathogen Providencia alcalifaciens N-formyltransferase ankyrin repeats function in small molecule binding and allosteric control. Ankyrin-repeat proteins have been associated with a number of human diseases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.90e-239 | 1 | 716 | 3 | 793 | |
| 8.88e-144 | 49 | 374 | 50 | 373 | |
| 2.18e-141 | 1 | 374 | 5 | 382 | |
| 2.18e-141 | 1 | 374 | 5 | 382 | |
| 2.18e-141 | 1 | 374 | 5 | 382 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.09e-29 | 415 | 715 | 4 | 291 | Solute carrier family 35 member E3 OS=Mus musculus OX=10090 GN=Slc35e3 PE=2 SV=1 |
|
| 7.68e-29 | 436 | 715 | 20 | 291 | Solute carrier family 35 member E3 OS=Bos taurus OX=9913 GN=SLC35E3 PE=2 SV=1 |
|
| 4.74e-28 | 429 | 715 | 13 | 291 | Solute carrier family 35 member E3 OS=Danio rerio OX=7955 GN=slc35e3 PE=2 SV=1 |
|
| 4.74e-28 | 423 | 715 | 13 | 291 | Solute carrier family 35 member E3 OS=Homo sapiens OX=9606 GN=SLC35E3 PE=1 SV=1 |
|
| 2.22e-21 | 435 | 724 | 20 | 303 | UDP-rhamnose/UDP-galactose transporter 2 OS=Arabidopsis thaliana OX=3702 GN=URGT2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.992141 | 0.007887 |
TMHMM Annotations download full data without filtering help
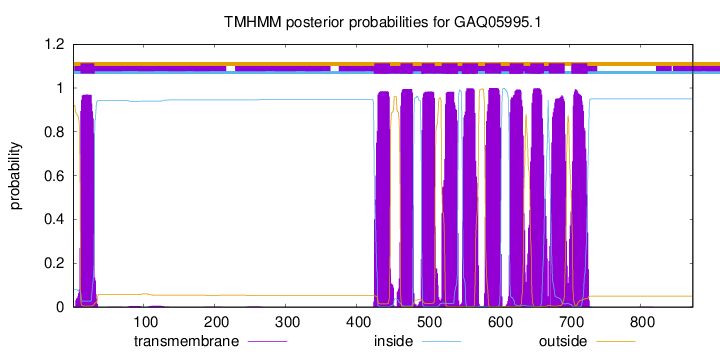
| Start | End |
|---|---|
| 11 | 30 |
| 425 | 447 |
| 462 | 479 |
| 492 | 510 |
| 520 | 542 |
| 549 | 571 |
| 581 | 603 |
| 616 | 635 |
| 645 | 664 |
| 671 | 693 |
| 703 | 725 |
