You are browsing environment: FUNGIDB
CAZyme Information: GAQ04950.1
You are here: Home > Sequence: GAQ04950.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ04950.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | xyloglucan-specific endo-beta-1,4-glucanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 182 | 335 | 1.4e-23 | 0.8974358974358975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 1.72e-09 | 88 | 337 | 3 | 207 | Glycosyl hydrolase family 12. |
| 238620 | AdeC | 0.006 | 142 | 247 | 45 | 137 | Adenine deaminase (AdeC) directly deaminates adenine to form hypoxanthine. This reaction is part of one of the adenine salvage pathways, as well as the degradation pathway. It is important for adenine utilization as a purine, as well as a nitrogen source in bacteria and archea. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.93e-109 | 1 | 337 | 1 | 312 | |
| 8.93e-109 | 1 | 337 | 1 | 312 | |
| 8.93e-109 | 1 | 337 | 1 | 312 | |
| 8.93e-109 | 1 | 337 | 1 | 312 | |
| 1.18e-101 | 7 | 337 | 9 | 333 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.46e-11 | 85 | 334 | 19 | 226 | The X-ray Crystal Structure of the Trichoderma harzianum Endoglucanase 3 from family GH12 [Trichoderma harzianum],4H7M_B The X-ray Crystal Structure of the Trichoderma harzianum Endoglucanase 3 from family GH12 [Trichoderma harzianum] |
|
| 5.53e-09 | 88 | 320 | 16 | 203 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
|
| 6.08e-08 | 88 | 320 | 15 | 202 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
|
| 5.63e-06 | 88 | 320 | 15 | 207 | The structure of Aspergillus niger endoglucanase-palladium complex [Aspergillus niger],1KS5_A Structure of Aspergillus niger endoglucanase [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.68e-08 | 88 | 322 | 36 | 227 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger OX=5061 GN=xgeA PE=1 SV=1 |
|
| 2.68e-08 | 88 | 322 | 36 | 227 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xgeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
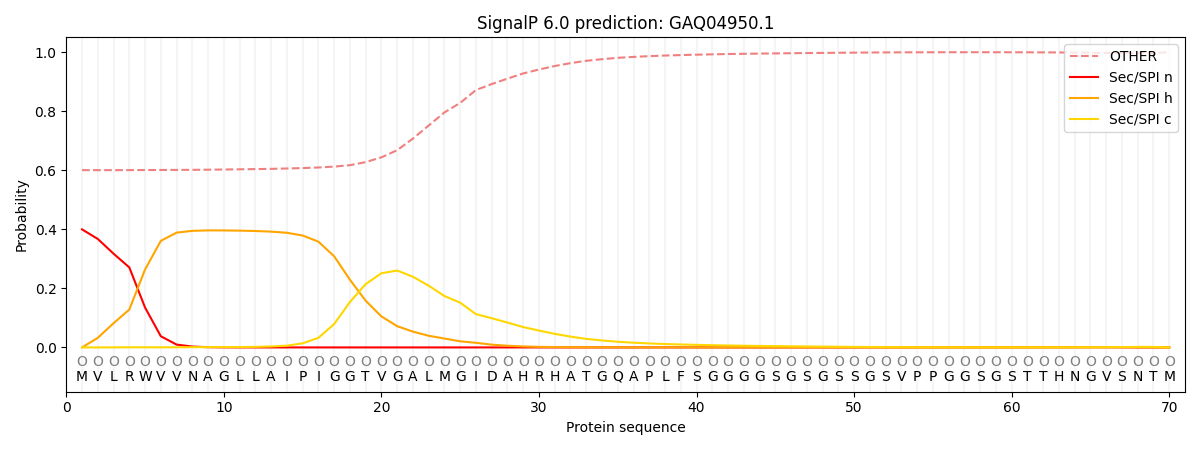
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.620769 | 0.379226 |

