You are browsing environment: FUNGIDB
CAZyme Information: GAQ04921.1
You are here: Home > Sequence: GAQ04921.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus lentulus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus lentulus | |||||||||||
| CAZyme ID | GAQ04921.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | pdp3-interacting factor 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 30 | 206 | 5.8e-67 | 0.9162303664921466 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226802 | MtnX | 1.27e-54 | 203 | 428 | 5 | 217 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase (methionine salvage) [Amino acid transport and metabolism]. |
| 319826 | HAD_Pase | 4.13e-49 | 203 | 423 | 1 | 210 | phosphatase, similar to Bacillus subtilis MtnX; belongs to the haloacid dehalogenase-like superfamily. Bacillus subtilis recycles two toxic byproducts of polyamine metabolism, methylthioadenosine and methylthioribose, into methionine by a salvage pathway. The sixth reaction in this pathway is catalyzed by B. subtilis MtnX: the dephosphorylation of 2- hydroxy-3-keto-5-methylthiopentenyl-1-phosphate (HKMTP- 1-P) into 1,2-dihydroxy-3-keto-5-methylthiopentene. The hydrolysis of HK-MTP-1-P is a two-step mechanism involving the formation of a transiently phosphorylated aspartyl intermediate. Members of this family belong to the haloacid dehalogenase-like (HAD) hydrolases, a large superfamily of diverse enzymes that catalyze carbon or phosphoryl group transfer reactions on a range of substrates, using an active site aspartate in nucleophilic catalysis. Members of this superfamily include 2-L-haloalkanoic acid dehalogenase, azetidine hydrolase, phosphonoacetaldehyde hydrolase, phosphoserine phosphatase, phosphomannomutase, P-type ATPases and many others. HAD hydrolases are found in all three kingdoms of life, and most genomes are predicted to contain multiple HAD-like proteins. Members possess a highly conserved alpha/beta core domain, and many also possess a small cap domain, the fold and function of which is variable. HAD hydrolases are sometimes referred to as belonging to the DDDD superfamily of phosphohydrolases. |
| 213629 | DKMTPPase-SF | 2.29e-26 | 203 | 394 | 3 | 188 | 2,3-diketo-5-methylthio-1-phosphopentane phosphatase. This phosphatase is a member of the IB subfamily (TIGR01488) of the haloacid dehalogenase (HAD) superfamily of aspartate-nucleophile hydrolases. With the exception of OMNI|NTL01BS01361 from B. subtilis and GP|15024582 from Clostridium acetabutylicum, the members of this group are all eukaryotic, spanning metazoa, plants and fungi. The B. subtilus gene (YkrX, renamed MtnX) is part of an operon for the conversion of methylthioribose (MTR) to methionine. It works with the enolase MtnW, a RuBisCO homolog. The combination of MtnW and MtnX achieves the same overall reaction as the enolase-phosphatase MtnC. The function of MtnX was shown by Ashida, et al. (2003) to be 2,3-diketo-5-methylthio-1-phosphopentane phosphatase, rather than 2,3-diketo-5-methylthio-1-phosphopentane phosphatase as proposed earlier. See the Genome Property for methionine salvage for more details. In eukaryotes, methionine salvage from methylthioadenosine also occurs. It seems reasonable that members of this family in eukaryotes fulfill a similar role as in Bacillus. A more specific, equivalog-level model is TIGR03333. Note that SP|P53981 from S. cerevisiae, a member of this family, is annotated as a "probable membrane protein" due to a predicted transmembrane helix. The region in question contains the second of the three conserved HAD superfamily catalytic motifs and thus, considering the fold of the HAD catalytic domain, is unlikely to be a transmembrane region in fact. [Central intermediary metabolism, Other] |
| 236562 | mtnX | 5.56e-19 | 203 | 425 | 5 | 215 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase; Reviewed |
| 273653 | HAD-SF-IB | 1.44e-15 | 207 | 387 | 5 | 177 | Haloacid Dehalogenase superfamily, subfamily IB, phosphoserine phosphatase-like. This model represents a subfamily of the Haloacid Dehalogenase superfamily of aspartate-nucleophile hydrolases. Subfamily IA, B, C and D are distinguished from the rest of the superfamily by the presence of a variable domain between the first and second conserved catalytic motifs. In subfamilies IA and IB, this domain consists of an alpha-helical bundle. It was necessary to model these two subfamilies separately, breaking them at a an apparent phylogenetic bifurcation, so that the resulting model(s) are not so broadly defined that members of subfamily III (which lack the variable domain) are included. Subfamily IA includes the enzyme phosphoserine phosphatase (TIGR00338) as well as three hypothetical equivalogs. Many members of these hypothetical equivalogs have been annotated as PSPase-like or PSPase-family proteins. In particular, the hypothetical equivalog which appears to be most closely related to PSPase contains only Archaea (while TIGR00338 contains only eukaryotes and bacteria) of which some are annotated as PSPases. Although this is a reasonable conjecture, none of these sequences has sufficient evidence for this assignment. If such should be found, this model should be retired while the PSPase model should be broadened to include these sequences. [Unknown function, Enzymes of unknown specificity] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.96e-106 | 4 | 206 | 6 | 208 | |
| 1.59e-98 | 11 | 205 | 10 | 204 | |
| 1.59e-98 | 11 | 205 | 10 | 204 | |
| 3.52e-98 | 3 | 207 | 5 | 209 | |
| 5.43e-98 | 11 | 205 | 46 | 240 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.50e-35 | 20 | 205 | 1 | 201 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
|
| 2.24e-08 | 270 | 431 | 74 | 223 | Crystal structure of MtnX phosphatase from Bacillus Subtilis at 2.00 A resolution [Bacillus subtilis],2FEA_B Crystal structure of MtnX phosphatase from Bacillus Subtilis at 2.00 A resolution [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.29e-49 | 204 | 425 | 9 | 225 | Pdp3-interacting factor 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ptf1 PE=1 SV=1 |
|
| 4.28e-42 | 203 | 426 | 5 | 229 | Polyol phosphate phosphatase PYP1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PYP1 PE=1 SV=1 |
|
| 1.41e-14 | 204 | 427 | 5 | 216 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase OS=Bacillus mycoides (strain KBAB4) OX=315730 GN=mtnX PE=3 SV=1 |
|
| 3.51e-14 | 204 | 427 | 5 | 216 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase OS=Bacillus cytotoxicus (strain DSM 22905 / CIP 110041 / 391-98 / NVH 391-98) OX=315749 GN=mtnX PE=3 SV=1 |
|
| 1.18e-13 | 262 | 424 | 63 | 214 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase OS=Exiguobacterium sibiricum (strain DSM 17290 / CIP 109462 / JCM 13490 / 255-15) OX=262543 GN=mtnX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
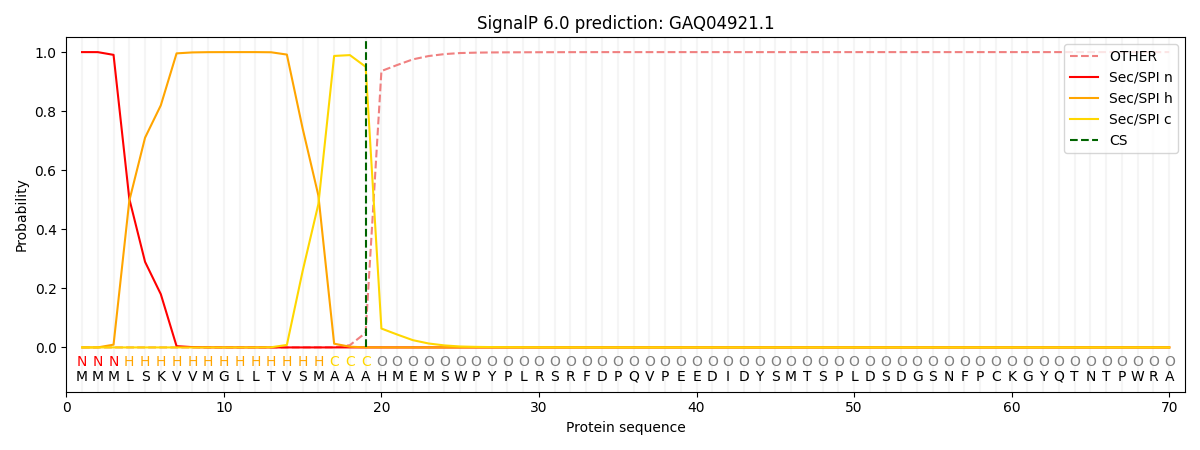
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000226 | 0.999755 | CS pos: 19-20. Pr: 0.9504 |
