You are browsing environment: FUNGIDB
CAZyme Information: FUN_022248-T1-p1
You are here: Home > Sequence: FUN_022248-T1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | FUN_022248-T1-p1 | |||||||||||
| CAZy Family | GT33 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT5 | 112 | 680 | 1.9e-40 | 0.9936440677966102 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223374 | GlgA | 1.68e-19 | 112 | 589 | 3 | 398 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 340822 | GT5_Glycogen_synthase_DULL1-like | 2.72e-19 | 112 | 589 | 2 | 398 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 400563 | Glyco_transf_5 | 3.40e-14 | 112 | 321 | 1 | 166 | Starch synthase catalytic domain. |
| 234809 | glgA | 1.83e-11 | 128 | 589 | 20 | 386 | glycogen synthase GlgA. |
| 172588 | PRK14098 | 2.45e-09 | 284 | 575 | 143 | 397 | starch synthase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.01e-106 | 85 | 683 | 127 | 802 | |
| 5.96e-103 | 47 | 682 | 63 | 719 | |
| 1.71e-96 | 83 | 691 | 111 | 813 | |
| 6.11e-18 | 112 | 683 | 191 | 715 | |
| 9.21e-13 | 112 | 613 | 36 | 482 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.38e-09 | 254 | 679 | 114 | 485 | Glycogen synthase 1 OS=Synechococcus sp. (strain JA-2-3B'a(2-13)) OX=321332 GN=glgA1 PE=3 SV=1 |
|
| 2.45e-09 | 235 | 689 | 108 | 493 | Glycogen synthase OS=Thermodesulfovibrio yellowstonii (strain ATCC 51303 / DSM 11347 / YP87) OX=289376 GN=glgA PE=3 SV=1 |
|
| 2.96e-08 | 254 | 679 | 114 | 485 | Glycogen synthase 1 OS=Synechococcus sp. (strain JA-3-3Ab) OX=321327 GN=glgA1 PE=3 SV=1 |
|
| 3.60e-07 | 112 | 575 | 8 | 397 | Glycogen synthase OS=Pelodictyon phaeoclathratiforme (strain DSM 5477 / BU-1) OX=324925 GN=glgA PE=3 SV=1 |
|
| 1.09e-06 | 254 | 679 | 111 | 481 | Glycogen synthase 2 OS=Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath) OX=243233 GN=glgA2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
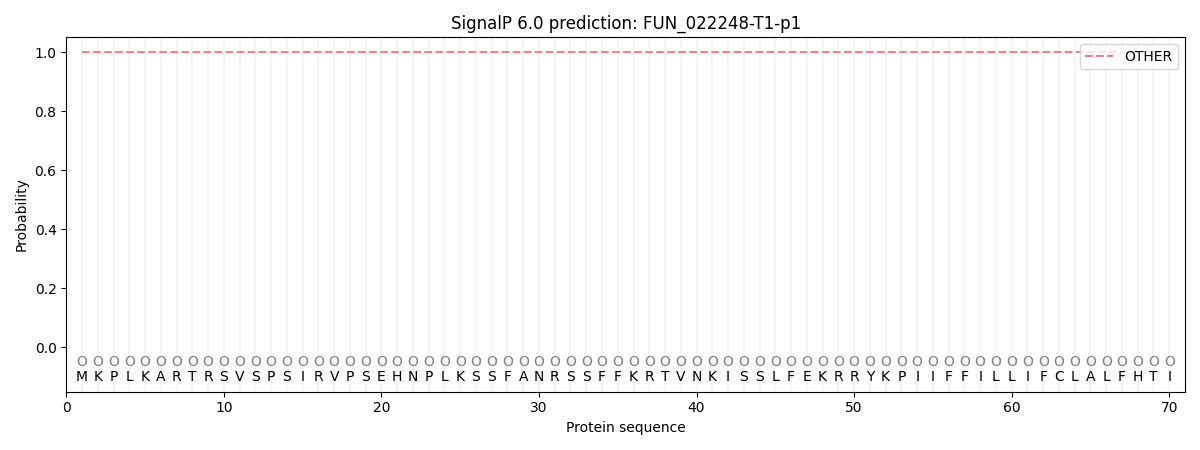
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999973 | 0.000050 |

