You are browsing environment: FUNGIDB
CAZyme Information: FUN_009493-T1-p1
You are here: Home > Sequence: FUN_009493-T1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | FUN_009493-T1-p1 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 90 | 558 | 7.6e-114 | 0.9776536312849162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 7.49e-88 | 73 | 563 | 1 | 520 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 215324 | PLN02604 | 8.32e-79 | 73 | 558 | 24 | 540 | oxidoreductase |
| 177843 | PLN02191 | 9.32e-76 | 86 | 583 | 36 | 563 | L-ascorbate oxidase |
| 225043 | SufI | 8.13e-74 | 61 | 563 | 20 | 447 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| 274556 | laccase | 3.95e-65 | 92 | 575 | 22 | 530 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.39e-109 | 73 | 585 | 122 | 658 | |
| 3.58e-108 | 68 | 582 | 181 | 722 | |
| 1.65e-106 | 74 | 571 | 23 | 503 | |
| 6.14e-105 | 68 | 582 | 62 | 591 | |
| 8.64e-105 | 68 | 582 | 62 | 591 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-98 | 78 | 586 | 8 | 490 | Laccase from Antrodiella faginea [Antrodiella faginea] |
|
| 5.66e-97 | 59 | 586 | 10 | 511 | Crystal structure of laccases from Pycnoporus sanguineus, izoform I [Trametes sanguinea] |
|
| 1.32e-94 | 59 | 586 | 10 | 511 | Crystal structure of laccases from Pycnoporus sanguineus, izoform II [Trametes coccinea],5NQ9_A Crystal structure of laccases from Pycnoporus sanguineus, izoform II, monoclinic [Trametes sanguinea],5NQ9_C Crystal structure of laccases from Pycnoporus sanguineus, izoform II, monoclinic [Trametes sanguinea] |
|
| 2.70e-94 | 61 | 582 | 4 | 484 | Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena],5Z1X_B Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena],5Z22_A Crystal Structure of Laccase from Cerrena sp. RSD1 [Cerrena] |
|
| 6.31e-93 | 61 | 586 | 5 | 492 | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Third structure of the series with 315 KGy dose. [Steccherinum murashkinskyi],6RHI_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. [Steccherinum murashkinskyi],6RHO_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Twentieth structure of the series with 4065 KGy dose. [Steccherinum murashkinskyi],6RHP_A Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Twenty first structure of the series with 4415 KGy dose (collected after refreezing). [Steccherinum murashkinskyi],6RHR_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. First structure of the series with 15 KGy dose. [Steccherinum murashkinskyi],6RHU_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Second structure of the series with 165 KGy dose. [Steccherinum murashkinskyi],6RHX_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Third structure of the series with 315 KGy dose. [Steccherinum murashkinskyi],6RI0_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Ninth structure of the series with 1215 KGy dose. [Steccherinum murashkinskyi],6RI2_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Twentieth structure of the series with 4065 KGy dose. [Steccherinum murashkinskyi],6RI4_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. First structure of the series with 13 KGy dose. [Steccherinum murashkinskyi],6RI6_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Second structure of the series with 400 KGy dose. [Steccherinum murashkinskyi],6RI8_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Third structure of the series with 800 KGy dose. [Steccherinum murashkinskyi],6RII_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourth structure of the series with 1200 KGy dose. [Steccherinum murashkinskyi],6RIK_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Thirteenth structure of the series with 5200 KGy dose. [Steccherinum murashkinskyi],6RIL_A Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourteenth structure of the series with 5600 KGy dose (data was collected after refreezing). [Steccherinum murashkinskyi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.27e-98 | 74 | 562 | 23 | 494 | Laccase-4 OS=Thanatephorus cucumeris OX=107832 GN=LCC4 PE=1 SV=1 |
|
| 1.21e-95 | 71 | 582 | 25 | 509 | Laccase OS=Phlebia radiata OX=5308 GN=LAC PE=1 SV=2 |
|
| 4.84e-94 | 59 | 586 | 10 | 511 | Laccase OS=Pycnoporus cinnabarinus OX=5643 GN=LCC3-1 PE=1 SV=1 |
|
| 4.27e-92 | 78 | 586 | 29 | 513 | Laccase OS=Trametes hirsuta OX=5327 PE=1 SV=1 |
|
| 1.38e-89 | 73 | 586 | 21 | 513 | Laccase-1 OS=Agaricus bisporus OX=5341 GN=lcc1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
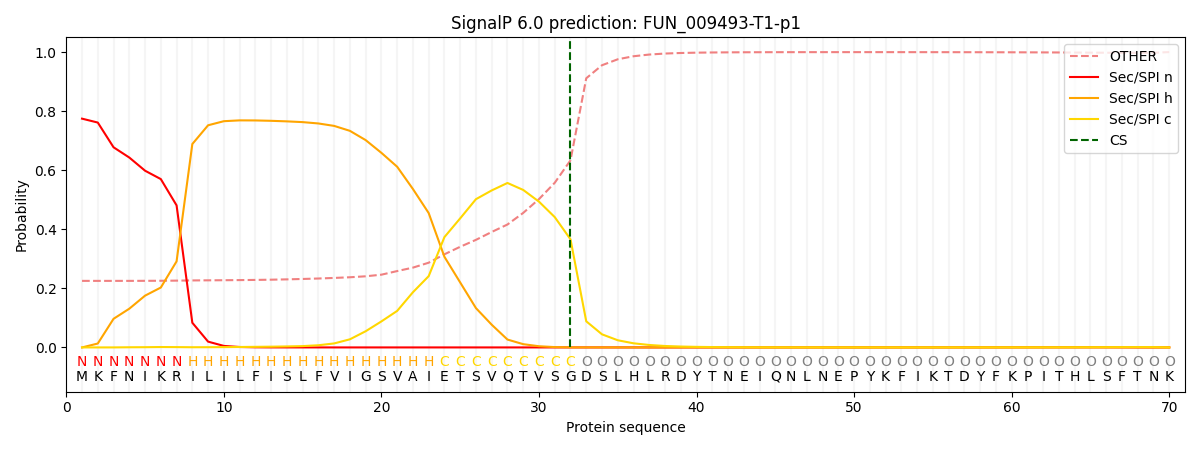
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.238188 | 0.761788 | CS pos: 32-33. Pr: 0.3661 |

