You are browsing environment: FUNGIDB
CAZyme Information: FUN_004908-T1-p1
You are here: Home > Sequence: FUN_004908-T1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
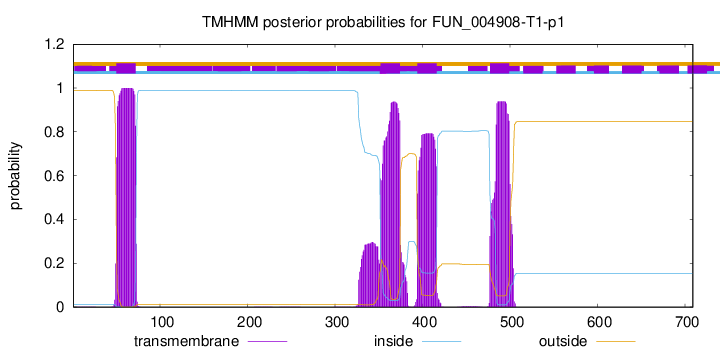
TMHMM annotations
Basic Information help
| Species | Rhizophagus irregularis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Glomeromycetes; ; Glomeraceae; Rhizophagus; Rhizophagus irregularis | |||||||||||
| CAZyme ID | FUN_004908-T1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT21 | 94 | 325 | 2.8e-95 | 0.9957081545064378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133012 | Glucosylceramide_synthase | 2.33e-86 | 94 | 325 | 1 | 196 | Glucosylceramide synthase catalyzes the first glycosylation step of glycosphingolipid synthesis. UDP-glucose:N-acylsphingosine D-glucosyltransferase (glucosylceramide synthase or ceramide glucosyltransferase) catalyzes the first glycosylation step of glycosphingolipid synthesis. Its product, glucosylceramide, serves as the core of more than 300 glycosphingolipids (GSL). GSLs are a group of membrane components that have the lipid portion embedded in the outer plasma membrane leaflet and the sugar chains extended to the outer environment. Several lines of evidence suggest the importance of GSLs in various cellular processes such as differentiation, adhesion, proliferation, and cell-cell recognition. In pathogenic fungus Cryptococcus neoformans, glucosylceramide serves as an antigen that elicits an antibody response in patients and it is essential for fungal growth in host extracellular environment. |
| 132512 | HpnI | 3.08e-44 | 94 | 437 | 41 | 373 | hopanoid biosynthesis associated glycosyl transferase protein HpnI. This family of genes include a glycosyl transferase, group 2 domain (pfam00535) which are responsible, generally for the transfer of nucleotide-diphosphate sugars to substrates such as polysaccharides and lipids. The member of this clade from Acidithiobacillus ferrooxidans ATCC 23270 (AFE_0974) is found in the same locus as squalene-hopene cyclase (SHC, TIGR01507) and other genes associated with the biosynthesis of hopanoid natural products. Similarly, in Ralstonia eutropha JMP134 (Reut_B4902) this gene is adjacent to HpnAB, IspH and HpnH (TIGR03470), although SHC itself is elsewhere in the genome. Notably, this gene (here named HpnI) and three others form a conserved set (HpnIJKL) which occur in a subset of all genomes containing the SHC enzyme. This relationship was discerned using the method of partial phylogenetic profiling. This group includes Zymomonas mobilis, the organism where the initial hopanoid biosynthesis locus was described consisting of the genes HpnA-E and SHC (HpnF). Continuing past SHC are found a phosphorylase enzyme (ZMO0873, i.e. HpnG, TIGR03468) and another radical SAM enzyme (ZMO0874), HpnH. Although discontinuous in Z. mobilis, we continue the gene symbol sequence with HpnIJKL. Hopanoids are known to feature polar glycosyl head groups in many organisms. |
| 404400 | Glyco_transf_21 | 1.08e-40 | 149 | 323 | 1 | 174 | Glycosyl transferase family 21. This is a family of ceramide beta-glucosyltransferases - EC:2.4.1.80. |
| 224136 | BcsA | 7.80e-20 | 49 | 430 | 15 | 428 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 404520 | Glyco_tranf_2_3 | 1.85e-13 | 94 | 323 | 2 | 229 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.76e-133 | 66 | 441 | 27 | 412 | |
| 2.56e-108 | 74 | 450 | 24 | 428 | |
| 2.37e-106 | 90 | 450 | 47 | 445 | |
| 2.47e-97 | 93 | 437 | 51 | 429 | |
| 3.00e-95 | 67 | 439 | 37 | 447 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.94e-93 | 53 | 441 | 13 | 450 | Ceramide glucosyltransferase OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=GCS1 PE=1 SV=1 |
|
| 2.07e-74 | 49 | 437 | 13 | 391 | Ceramide glucosyltransferase OS=Rattus norvegicus OX=10116 GN=Ugcg PE=1 SV=1 |
|
| 4.48e-74 | 92 | 423 | 80 | 495 | Ceramide glucosyltransferase OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=PAS_chr3_0357 PE=1 SV=1 |
|
| 5.66e-74 | 49 | 437 | 13 | 391 | Ceramide glucosyltransferase OS=Mus musculus OX=10090 GN=Ugcg PE=1 SV=1 |
|
| 3.02e-73 | 53 | 437 | 17 | 391 | Ceramide glucosyltransferase OS=Homo sapiens OX=9606 GN=UGCG PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999874 | 0.000151 |