You are browsing environment: FUNGIDB
CAZyme Information: FPRO_15743-t41_1-p1
You are here: Home > Sequence: FPRO_15743-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_15743-t41_1-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | related to long chain fatty alcohol oxidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 192 | 705 | 8.7e-66 | 0.9671532846715328 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 366272 | GMC_oxred_N | 6.85e-63 | 244 | 466 | 1 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 225186 | BetA | 1.37e-37 | 193 | 720 | 4 | 542 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 4.53e-26 | 545 | 703 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 235000 | PRK02106 | 4.07e-07 | 598 | 704 | 438 | 528 | choline dehydrogenase; Validated |
| 274143 | pyranose_ox | 1.55e-04 | 642 | 698 | 477 | 533 | pyranose oxidase. Pyranose oxidase (also called glucose 2-oxidase) converts D-glucose and molecular oxygen to 2-dehydro-D-glucose and hydrogen peroxide. Peroxide production is believed to be important to the wood rot fungi in which this enzyme is found for lignin degradation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 721 | 8 | 744 | |
| 7.39e-15 | 198 | 711 | 7 | 596 | |
| 2.20e-14 | 193 | 711 | 2 | 597 | |
| 3.84e-14 | 193 | 711 | 2 | 597 | |
| 1.49e-13 | 198 | 704 | 255 | 783 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.51e-08 | 198 | 714 | 231 | 770 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 3.95e-06 | 376 | 710 | 218 | 567 | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase [Aspergillus flavus NRRL3357],4YNU_A Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone [Aspergillus flavus NRRL3357] |
|
| 6.57e-06 | 246 | 714 | 98 | 511 | Chain A, 6'''-hydroxyparomomycin C oxidase [Microbacterium trichothecenolyticum],7DVE_B Chain B, 6'''-hydroxyparomomycin C oxidase [Microbacterium trichothecenolyticum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.35e-90 | 194 | 714 | 234 | 747 | Long-chain-alcohol oxidase FAO1 OS=Lotus japonicus OX=34305 GN=FAO1 PE=1 SV=1 |
|
| 1.45e-88 | 194 | 713 | 236 | 748 | Long-chain-alcohol oxidase FAO2 OS=Lotus japonicus OX=34305 GN=FAO2 PE=2 SV=1 |
|
| 1.03e-83 | 194 | 711 | 230 | 740 | Long-chain-alcohol oxidase FAO3 OS=Arabidopsis thaliana OX=3702 GN=FAO3 PE=1 SV=1 |
|
| 5.62e-81 | 187 | 716 | 228 | 747 | Long-chain-alcohol oxidase FAO4B OS=Arabidopsis thaliana OX=3702 GN=FAO4B PE=2 SV=2 |
|
| 9.74e-80 | 111 | 713 | 88 | 724 | Long-chain-alcohol oxidase FAO4A OS=Arabidopsis thaliana OX=3702 GN=FAO4A PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
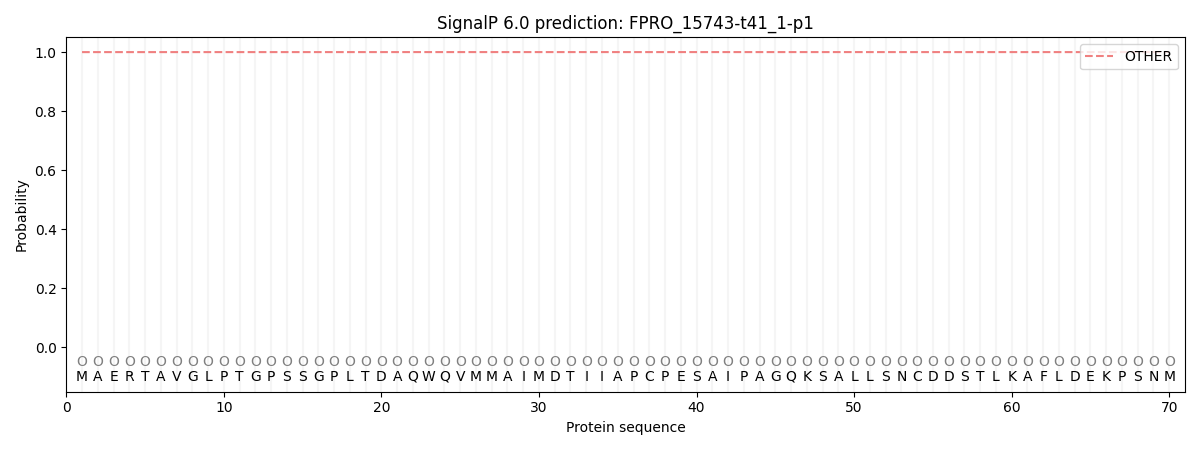
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000034 | 0.000000 |
