You are browsing environment: FUNGIDB
CAZyme Information: FPRO_14923-t41_1-p1
You are here: Home > Sequence: FPRO_14923-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_14923-t41_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 20 | 555 | 3.2e-132 | 0.9848197343453511 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404168 | Beta_helix | 9.52e-05 | 334 | 459 | 1 | 118 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| 404168 | Beta_helix | 3.00e-04 | 375 | 565 | 7 | 141 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 743 | 1 | 717 | |
| 0.0 | 1 | 689 | 1 | 670 | |
| 0.0 | 1 | 743 | 1 | 673 | |
| 0.0 | 1 | 743 | 1 | 673 | |
| 0.0 | 1 | 743 | 1 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.16e-44 | 18 | 567 | 24 | 590 | Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
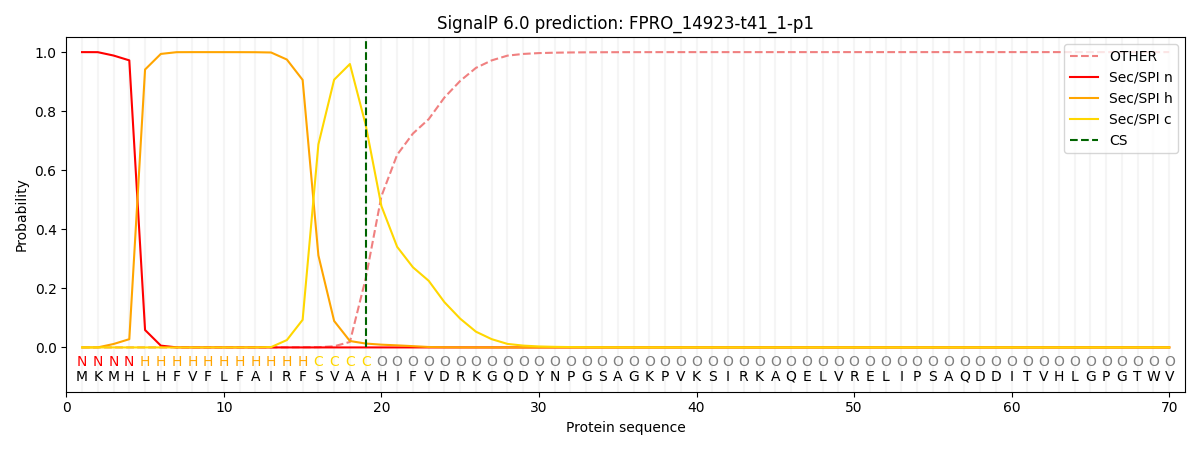
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000569 | 0.999395 | CS pos: 19-20. Pr: 0.7539 |
