You are browsing environment: FUNGIDB
CAZyme Information: FPRO_14119-t41_1-p1
You are here: Home > Sequence: FPRO_14119-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_14119-t41_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | probable alpha-glucuronidase precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.131:5 | 3.2.1.139:5 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH67 | 18 | 697 | 5.1e-269 | 0.9985052316890882 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226187 | AguA2 | 0.0 | 16 | 697 | 4 | 679 | Alpha-glucuronidase [Carbohydrate transport and metabolism]. |
| 400047 | Glyco_hydro_67M | 0.0 | 144 | 472 | 1 | 324 | Glycosyl hydrolase family 67 middle domain. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the central catalytic domain of alpha-glucuronidase. |
| 400039 | Glyco_hydro_67C | 1.28e-141 | 475 | 698 | 2 | 223 | Glycosyl hydrolase family 67 C-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the C terminal region of alpha-glucuronidase which is mainly alpha-helical. It wraps around the catalytic domain (pfam07488), making additional interactions both with the N-terminal domain (pfam03648) of its parent monomer and also forming the majority of the dimer-surface with the equivalent C-terminal domain of the other monomer of the dimer. |
| 397627 | Glyco_hydro_67N | 2.68e-28 | 22 | 141 | 1 | 120 | Glycosyl hydrolase family 67 N-terminus. Alpha-glucuronidases, components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans. This family represents the N-terminal region of alpha-glucuronidase. The N-terminal domain forms a two-layer sandwich, each layer being formed by a beta sheet of five strands. A further two helices form part of the interface with the central, catalytic, module (pfam07488). |
| 271143 | CBM6-CBM35-CBM36_like | 1.23e-10 | 704 | 835 | 1 | 124 | Carbohydrate Binding Module 6 (CBM6) and CBM35_like superfamily. Carbohydrate binding module family 6 (CBM6, family 6 CBM), also known as cellulose binding domain family VI (CBD VI), and related CBMs (CBM35 and CBM36). These are non-catalytic carbohydrate binding domains found in a range of enzymes that display activities against a diverse range of carbohydrate targets, including mannan, xylan, beta-glucans, cellulose, agarose, and arabinans. These domains facilitate the strong binding of the appended catalytic modules to their dedicated, insoluble substrates. Many of these CBMs are associated with glycoside hydrolase (GH) domains. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. CBM36s are calcium-dependent xylan binding domains. CBM35s display conserved specificity through extensive sequence similarity, but divergent function through their appended catalytic modules. This alignment model also contains the C-terminal domains of bacterial insecticidal toxins, where they may be involved in determining insect specificity through carbohydrate binding functionality. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 842 | 1 | 842 | |
| 0.0 | 1 | 842 | 1 | 842 | |
| 0.0 | 1 | 842 | 1 | 842 | |
| 0.0 | 1 | 842 | 1 | 842 | |
| 0.0 | 1 | 842 | 1 | 842 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.09e-189 | 23 | 697 | 9 | 675 | The Crystal Structure Of Alpha-D-Glucuronidase From Bacillus Stearothermophilus T-6 [Geobacillus stearothermophilus] |
|
| 7.19e-189 | 23 | 697 | 9 | 675 | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 [Geobacillus stearothermophilus],1L8N_A The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose [Geobacillus stearothermophilus],1MQQ_A THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID [Geobacillus stearothermophilus] |
|
| 1.44e-188 | 23 | 697 | 9 | 675 | Chain A, ALPHA-D-GLUCURONIDASE [Geobacillus stearothermophilus] |
|
| 2.86e-188 | 23 | 697 | 9 | 675 | Chain A, alpha-D-glucuronidase [Geobacillus stearothermophilus],1K9F_A Chain A, alpha-D-glucuronidase [Geobacillus stearothermophilus] |
|
| 2.90e-136 | 17 | 716 | 1 | 686 | Structure of Pseudomonas cellulosa alpha-D-glucuronidase [Cellvibrio japonicus],1GQI_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase [Cellvibrio japonicus],1GQJ_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose [Cellvibrio japonicus],1GQJ_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose [Cellvibrio japonicus],1GQK_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid [Cellvibrio japonicus],1GQK_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid [Cellvibrio japonicus],1GQL_A Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose [Cellvibrio japonicus],1GQL_B Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 15 | 842 | 16 | 847 | Alpha-glucuronidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aguA PE=1 SV=1 |
|
| 0.0 | 15 | 842 | 18 | 841 | Probable alpha-glucuronidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aguA PE=3 SV=1 |
|
| 0.0 | 16 | 842 | 18 | 835 | Probable alpha-glucuronidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aguA PE=3 SV=1 |
|
| 0.0 | 1 | 842 | 1 | 840 | Probable alpha-glucuronidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=aguA PE=3 SV=1 |
|
| 0.0 | 1 | 842 | 1 | 840 | Probable alpha-glucuronidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=aguA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
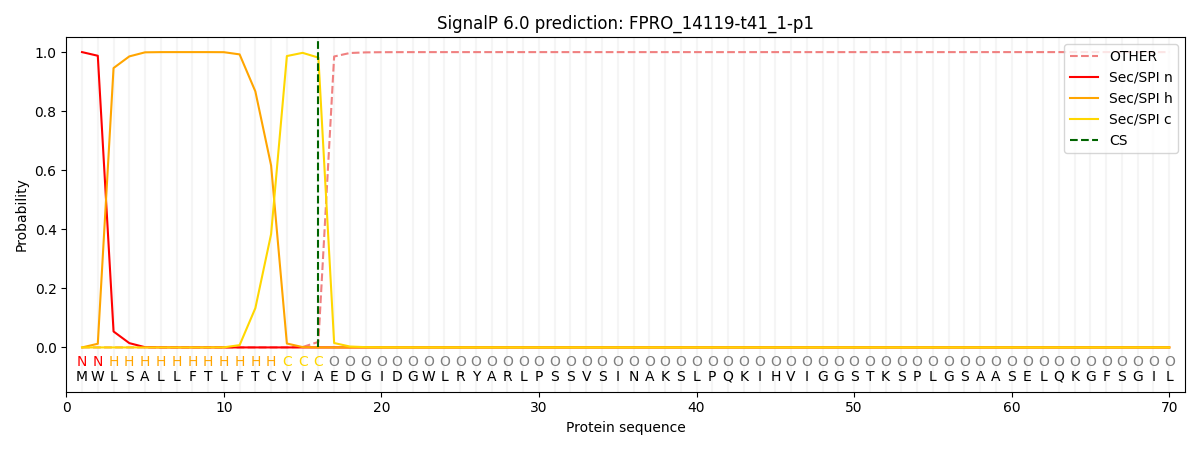
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000306 | 0.999667 | CS pos: 16-17. Pr: 0.9814 |
