You are browsing environment: FUNGIDB
CAZyme Information: FPRO_12535-t41_1-p1
You are here: Home > Sequence: FPRO_12535-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_12535-t41_1-p1 | |||||||||||
| CAZy Family | GH43|CBM91 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.74:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 25 | 219 | 1.4e-31 | 0.9947089947089947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.20e-20 | 26 | 219 | 2 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.13e-159 | 1 | 221 | 1 | 221 | |
| 8.13e-159 | 1 | 221 | 1 | 221 | |
| 8.13e-159 | 1 | 221 | 1 | 221 | |
| 2.72e-157 | 1 | 221 | 1 | 221 | |
| 8.39e-152 | 1 | 221 | 1 | 221 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.42e-94 | 23 | 221 | 3 | 203 | A novel cutinase-like protein from Cryptococcus sp. [Cryptococcus sp. S-2],2CZQ_B A novel cutinase-like protein from Cryptococcus sp. [Cryptococcus sp. S-2] |
|
| 1.53e-85 | 24 | 221 | 1 | 198 | Chain A, Cutinase-like enzyme [Moesziomyces antarcticus],7CC4_B Chain B, Cutinase-like enzyme [Moesziomyces antarcticus],7CW1_A Chain A, Cutinase-like enzyme [Moesziomyces antarcticus],7CW1_B Chain B, Cutinase-like enzyme [Moesziomyces antarcticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.94e-07 | 68 | 219 | 76 | 206 | Cutinase OS=Alternaria brassicicola OX=29001 GN=CUTAB1 PE=2 SV=1 |
|
| 5.04e-07 | 22 | 219 | 32 | 211 | Probable cutinase 1 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_084890 PE=3 SV=1 |
|
| 1.74e-06 | 22 | 219 | 32 | 211 | Probable cutinase 1 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_025250 PE=3 SV=1 |
|
| 1.74e-06 | 22 | 219 | 32 | 211 | Probable cutinase 1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_2G09380 PE=3 SV=1 |
|
| 6.89e-06 | 23 | 207 | 33 | 207 | Cutinase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=cut PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
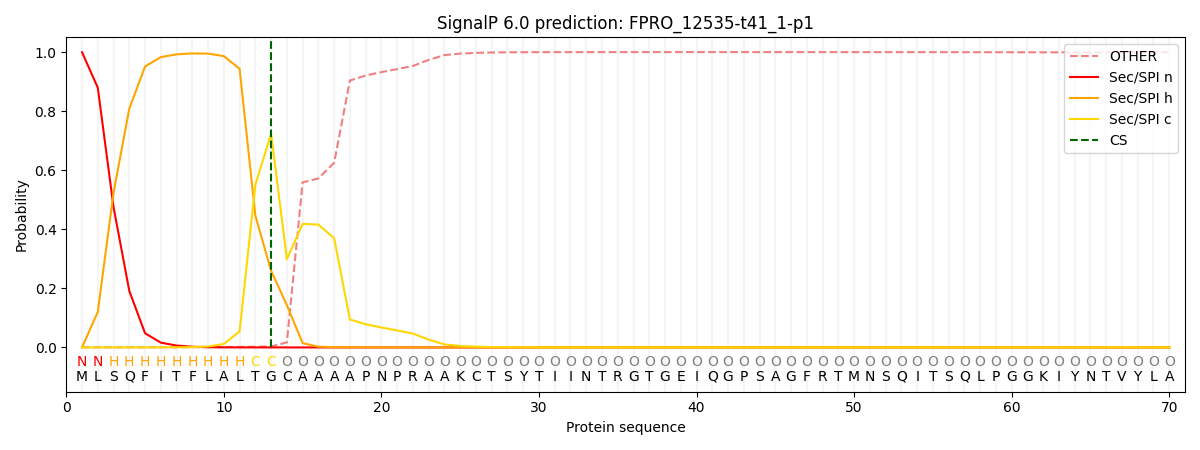
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001165 | 0.998839 | CS pos: 13-14. Pr: 0.7238 |
