You are browsing environment: FUNGIDB
CAZyme Information: FPRO_12447-t41_1-p1
You are here: Home > Sequence: FPRO_12447-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
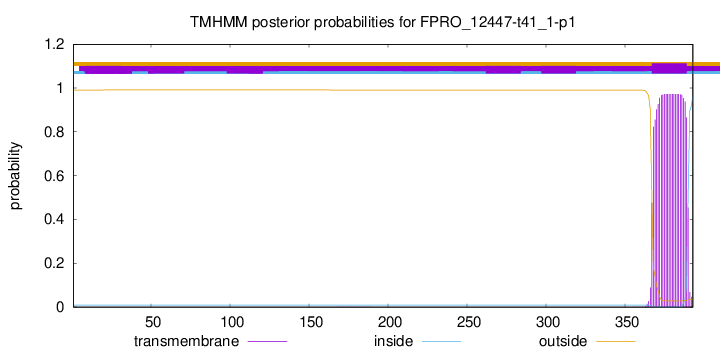
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_12447-t41_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | related to xyloglucan endo-transglycosylase-like protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 92 | 307 | 1.1e-90 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185683 | Glyco_hydrolase_16 | 2.01e-48 | 88 | 305 | 7 | 206 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.86e-301 | 1 | 393 | 1 | 393 | |
| 4.86e-301 | 1 | 393 | 1 | 393 | |
| 4.86e-301 | 1 | 393 | 1 | 393 | |
| 4.86e-301 | 1 | 393 | 1 | 393 | |
| 1.29e-278 | 1 | 393 | 1 | 394 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
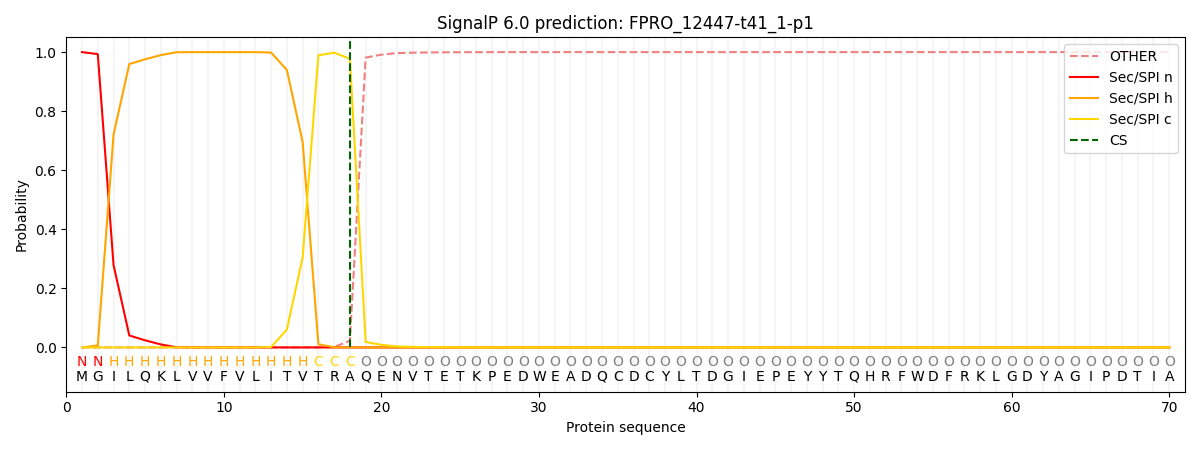
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000222 | 0.999748 | CS pos: 18-19. Pr: 0.9773 |