You are browsing environment: FUNGIDB
CAZyme Information: FPRO_12443-t41_1-p1
Basic Information
help
| Species |
Fusarium proliferatum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum
|
| CAZyme ID |
FPRO_12443-t41_1-p1
|
| CAZy Family |
GH43 |
| CAZyme Description |
uncharacterized protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 362 |
|
39401.06 |
7.2346 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FproliferatumET1 |
16509 |
1227346 |
366 |
16143
|
|
| Gene Location |
No EC number prediction in FPRO_12443-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
47 |
313 |
6.6e-45 |
0.9717314487632509 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 395098
|
Cellulase |
3.98e-05 |
86 |
311 |
57 |
268 |
Cellulase (glycosyl hydrolase family 5). |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.02e-277 |
1 |
362 |
1 |
362 |
| 3.02e-277 |
1 |
362 |
1 |
362 |
| 3.02e-277 |
1 |
362 |
1 |
362 |
| 3.02e-277 |
1 |
362 |
1 |
362 |
| 3.74e-262 |
1 |
362 |
1 |
362 |
FPRO_12443-t41_1-p1 has no PDB hit.
FPRO_12443-t41_1-p1 has no Swissprot hit.
This protein is predicted as SP
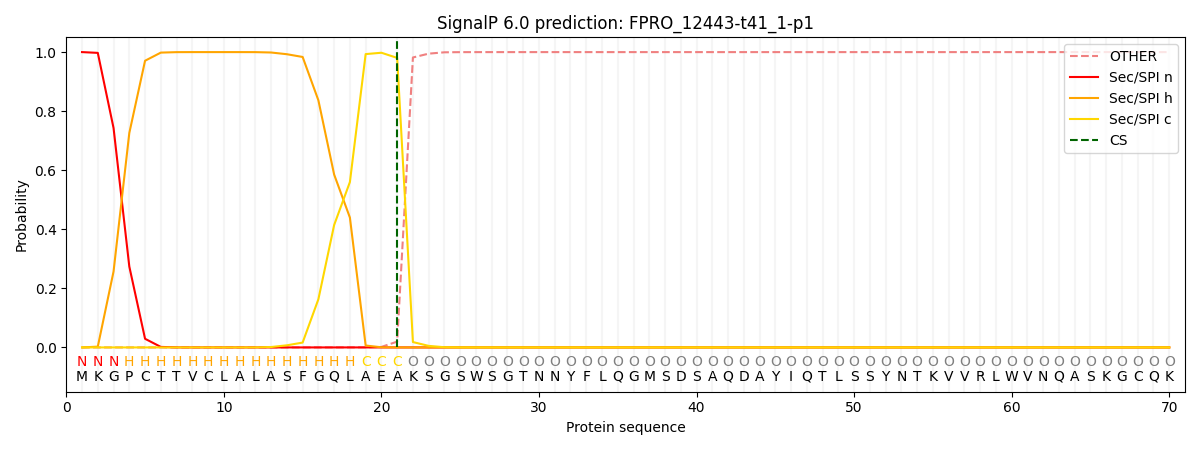
| Other |
SP_Sec_SPI |
CS Position |
| 0.000229 |
0.999758 |
CS pos: 21-22. Pr: 0.9804 |
There is no transmembrane helices in FPRO_12443-t41_1-p1.

