You are browsing environment: FUNGIDB
CAZyme Information: FPRO_07206-t41_1-p1
Basic Information
help
| Species |
Fusarium proliferatum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum
|
| CAZyme ID |
FPRO_07206-t41_1-p1
|
| CAZy Family |
GH13 |
| CAZyme Description |
uncharacterized protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 637 |
FJOF01000003|CGC2 |
72312.42 |
5.0894 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FproliferatumET1 |
16509 |
1227346 |
366 |
16143
|
|
| Gene Location |
No EC number prediction in FPRO_07206-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH142 |
11 |
465 |
3.3e-163 |
0.9603340292275574 |
FPRO_07206-t41_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
637 |
10 |
665 |
| 0.0 |
2 |
634 |
21 |
673 |
| 0.0 |
2 |
634 |
21 |
673 |
| 0.0 |
2 |
634 |
21 |
673 |
| 0.0 |
2 |
636 |
10 |
657 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.34e-90 |
16 |
465 |
653 |
1098 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 4.25e-87 |
16 |
465 |
653 |
1098 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 8.16e-16 |
16 |
459 |
305 |
800 |
Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.24e-23 |
16 |
624 |
336 |
1040 |
Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
This protein is predicted as OTHER
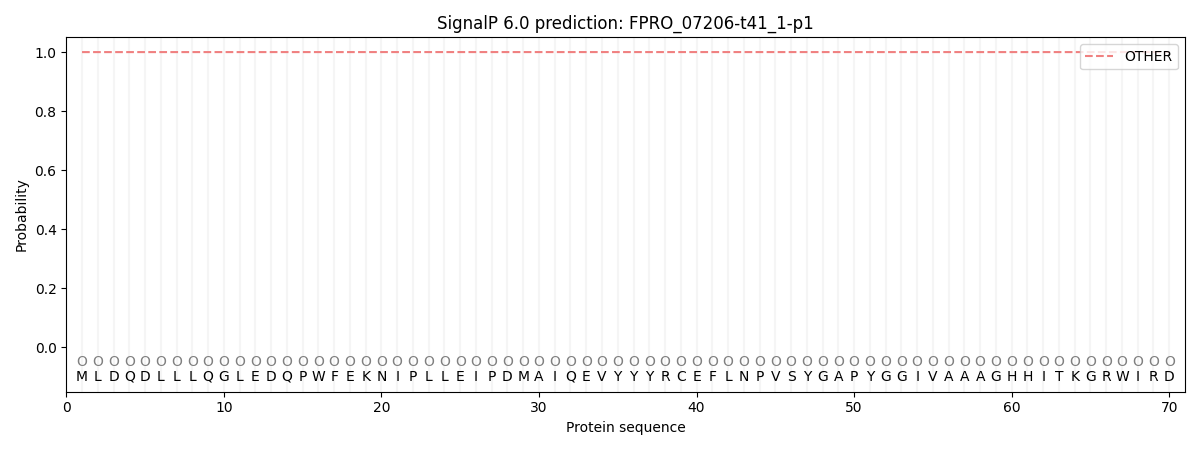
| Other |
SP_Sec_SPI |
CS Position |
| 1.000060 |
0.000000 |
|
There is no transmembrane helices in FPRO_07206-t41_1-p1.

