You are browsing environment: FUNGIDB
CAZyme Information: FPRO_06613-t41_1-p1
You are here: Home > Sequence: FPRO_06613-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_06613-t41_1-p1 | |||||||||||
| CAZy Family | CE8|CE8|CE8|CE8|CE8 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 483 | 681 | 4.4e-48 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 2.35e-50 | 483 | 681 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 238861 | SEST_like | 1.19e-30 | 52 | 329 | 1 | 238 | SEST_like. A family of secreted SGNH-hydrolases similar to Streptomyces scabies esterase (SEST), a causal agent of the potato scab disease, which hydrolyzes a specific ester bond in suberin, a plant lipid. The tertiary fold of this enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles two of the three components of typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxylic acid. |
| 404371 | Lipase_GDSL_2 | 8.60e-16 | 488 | 672 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238141 | SGNH_hydrolase | 2.23e-11 | 485 | 680 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 395531 | Lipase_GDSL | 2.76e-08 | 485 | 677 | 1 | 224 | GDSL-like Lipase/Acylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1436 | 1 | 1436 | |
| 0.0 | 1 | 1436 | 1 | 1437 | |
| 0.0 | 3 | 1436 | 5 | 1455 | |
| 0.0 | 451 | 1436 | 16 | 844 | |
| 0.0 | 466 | 1417 | 342 | 1309 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.24e-12 | 56 | 153 | 7 | 97 | Crystal structure of phospholipase A1 from Streptomyces albidoflavus NA297 [Streptomyces albidoflavus] |
|
| 7.50e-09 | 56 | 153 | 6 | 96 | Crystal structure of extracelular lipase from Streptomyces rimosus at 1.7A resolution [Streptomyces rimosus],5MAL_B Crystal structure of extracelular lipase from Streptomyces rimosus at 1.7A resolution [Streptomyces rimosus] |
|
| 4.53e-08 | 848 | 1046 | 422 | 606 | Crystal Structure of TcdB2-TccC3-Cdc42 [Photorhabdus luminescens] |
|
| 4.54e-08 | 848 | 1046 | 453 | 637 | Crystal Structure of TcdB2-TccC3 without hypervariable C-terminal region [Photorhabdus luminescens] |
|
| 4.55e-08 | 848 | 1046 | 487 | 671 | Crystal Structure of TcdB2-TccC3 [Photorhabdus luminescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.30e-09 | 56 | 153 | 40 | 130 | Lipase 1 OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO1725 PE=1 SV=1 |
|
| 5.57e-09 | 12 | 153 | 8 | 130 | Lipase OS=Streptomyces rimosus OX=1927 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
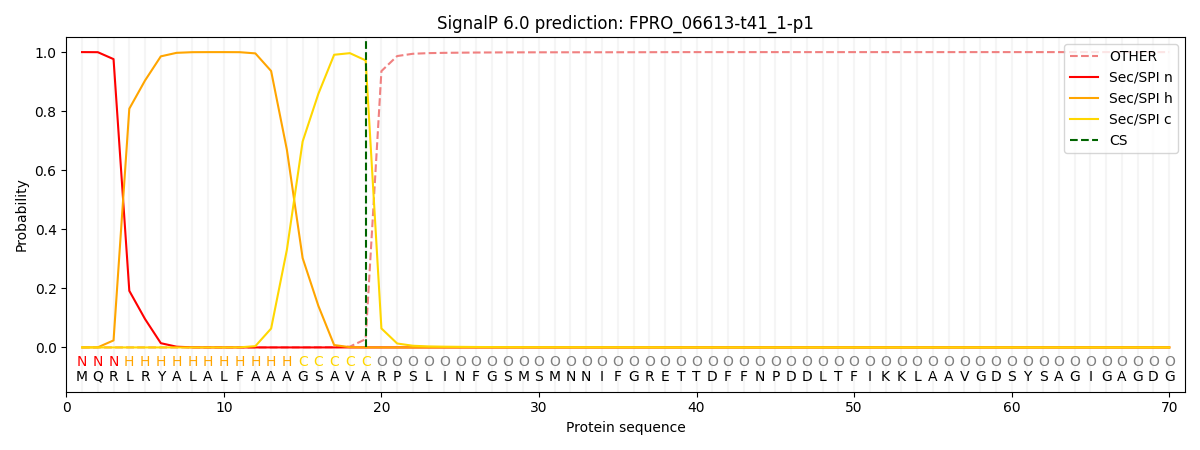
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000295 | 0.999677 | CS pos: 19-20. Pr: 0.9719 |
