You are browsing environment: FUNGIDB
CAZyme Information: FPRO_06138-t41_1-p1
Basic Information
help
| Species |
Fusarium proliferatum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum
|
| CAZyme ID |
FPRO_06138-t41_1-p1
|
| CAZy Family |
CE5 |
| CAZyme Description |
uncharacterized protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 662 |
FJOF01000003|CGC10 |
73817.25 |
5.7633 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FproliferatumET1 |
16509 |
1227346 |
366 |
16143
|
|
| Gene Location |
No EC number prediction in FPRO_06138-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH154 |
24 |
372 |
3.5e-123 |
0.9942528735632183 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 401852
|
DUF2264 |
5.76e-168 |
24 |
372 |
1 |
349 |
Uncharacterized protein conserved in bacteria (DUF2264). Members of this family of hypothetical bacterial proteins have no known function. |
| 226739
|
COG4289 |
6.12e-92 |
22 |
485 |
25 |
458 |
Uncharacterized protein [Function unknown]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
662 |
1 |
662 |
| 0.0 |
1 |
662 |
1 |
662 |
| 0.0 |
1 |
662 |
1 |
662 |
| 0.0 |
1 |
662 |
1 |
662 |
| 0.0 |
1 |
662 |
1 |
662 |
FPRO_06138-t41_1-p1 has no PDB hit.
FPRO_06138-t41_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
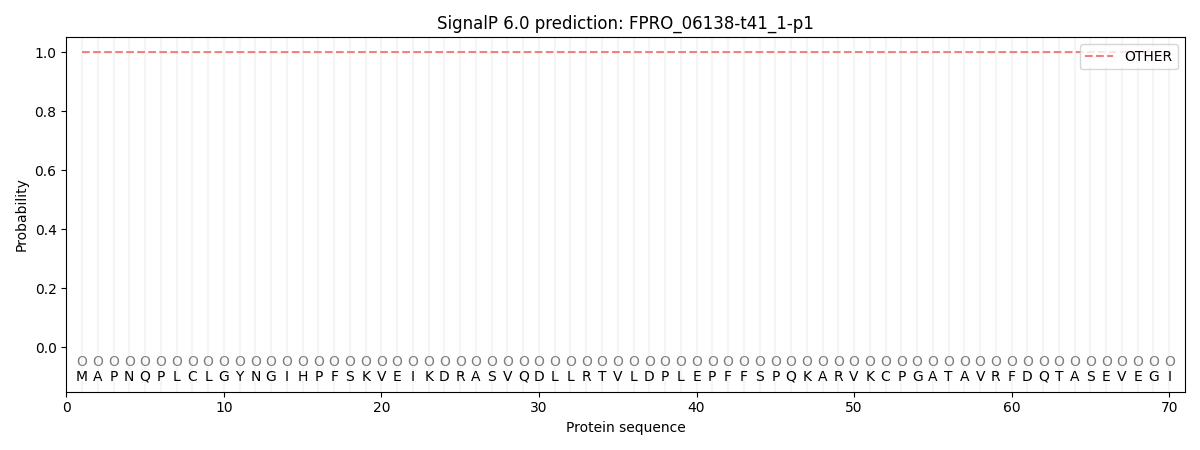
| Other |
SP_Sec_SPI |
CS Position |
| 1.000060 |
0.000000 |
|
There is no transmembrane helices in FPRO_06138-t41_1-p1.

