You are browsing environment: FUNGIDB
CAZyme Information: FPRO_03779-t41_1-p1
You are here: Home > Sequence: FPRO_03779-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_03779-t41_1-p1 | |||||||||||
| CAZy Family | AA8|AA3 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 131 | 410 | 2.2e-118 | 0.9964285714285714 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 0.010 | 68 | 213 | 7 | 153 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 437 | 1 | 437 | |
| 0.0 | 1 | 437 | 1 | 437 | |
| 0.0 | 1 | 437 | 1 | 437 | |
| 0.0 | 1 | 437 | 1 | 437 | |
| 0.0 | 1 | 437 | 1 | 437 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.46e-114 | 67 | 428 | 7 | 364 | Chain A, Glycoside hydrolase family 5 [Aspergillus oryzae],6LA0_B Chain B, Glycoside hydrolase family 5 [Aspergillus oryzae] |
|
| 9.76e-113 | 68 | 426 | 11 | 367 | Crystal structure of rutinosidase from Aspergillus niger [Aspergillus niger],6I1A_B Crystal structure of rutinosidase from Aspergillus niger [Aspergillus niger] |
|
| 7.73e-06 | 78 | 367 | 2 | 302 | Chain A, Exo-beta-1,3-glucanase [uncultured bacterium],6ZB9_B Chain B, Exo-beta-1,3-glucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.94e-19 | 72 | 362 | 35 | 310 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
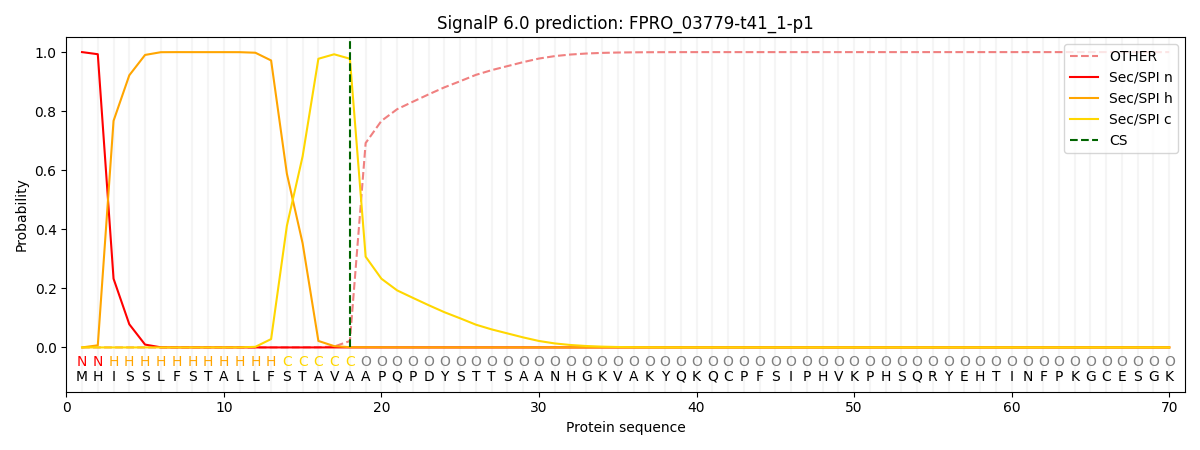
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000318 | 0.999660 | CS pos: 18-19. Pr: 0.9775 |
