You are browsing environment: FUNGIDB
CAZyme Information: FPRO_01997-t41_1-p1
You are here: Home > Sequence: FPRO_01997-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_01997-t41_1-p1 | |||||||||||
| CAZy Family | AA5 | |||||||||||
| CAZyme Description | related to 4-cresol dehydrogenase flavoprotein subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA4 | 26 | 546 | 5.8e-137 | 0.9808429118773946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 2.40e-27 | 79 | 214 | 5 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 2.17e-23 | 43 | 541 | 1 | 455 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 183043 | PRK11230 | 3.26e-07 | 81 | 274 | 62 | 254 | glycolate oxidase subunit GlcD; Provisional |
| 273751 | FAD_lactone_ox | 5.37e-06 | 80 | 211 | 20 | 147 | sugar 1,4-lactone oxidases. This model represents a family of at least two different sugar 1,4 lactone oxidases, both involved in synthesizing ascorbic acid or a derivative. These include L-gulonolactone oxidase (EC 1.1.3.8) from rat and D-arabinono-1,4-lactone oxidase (EC 1.1.3.37) from Saccharomyces cerevisiae. Members are proposed to have the cofactor FAD covalently bound at a site specified by Prosite motif PS00862; OX2_COVAL_FAD; 1. |
| 178402 | PLN02805 | 2.67e-05 | 38 | 275 | 96 | 325 | D-lactate dehydrogenase [cytochrome] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 556 | 1 | 556 | |
| 7.84e-161 | 5 | 545 | 7 | 556 | |
| 1.48e-159 | 5 | 544 | 10 | 559 | |
| 2.97e-159 | 5 | 547 | 10 | 562 | |
| 2.62e-153 | 5 | 545 | 6 | 555 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.02e-98 | 25 | 544 | 2 | 517 | Crystal structure of eugenol oxidase in complex with isoeugenol [Rhodococcus jostii RHA1],5FXD_B Crystal structure of eugenol oxidase in complex with isoeugenol [Rhodococcus jostii RHA1],5FXE_A Crystal structure of eugenol oxidase in complex with coniferyl alcohol [Rhodococcus jostii RHA1],5FXE_B Crystal structure of eugenol oxidase in complex with coniferyl alcohol [Rhodococcus jostii RHA1],5FXF_A Crystal structure of eugenol oxidase in complex with benzoate [Rhodococcus jostii RHA1],5FXF_B Crystal structure of eugenol oxidase in complex with benzoate [Rhodococcus jostii RHA1],5FXP_A Crystal structure of eugenol oxidase in complex with vanillin [Rhodococcus jostii RHA1],5FXP_B Crystal structure of eugenol oxidase in complex with vanillin [Rhodococcus jostii RHA1] |
|
| 3.85e-90 | 28 | 545 | 12 | 550 | STRUCTURE OF THE OCTAMERIC FLAVOENZYME VANILLYL-ALCOHOL OXIDASE: The505Ser Mutant [Penicillium simplicissimum],1W1J_B STRUCTURE OF THE OCTAMERIC FLAVOENZYME VANILLYL-ALCOHOL OXIDASE: The505Ser Mutant [Penicillium simplicissimum] |
|
| 3.85e-90 | 28 | 545 | 12 | 550 | STRUCTURE OF THE OCTAMERIC FLAVOENZYME VANILLYL-ALCOHOL OXIDASE: Ile238Thr Mutant [Penicillium simplicissimum],1W1K_B STRUCTURE OF THE OCTAMERIC FLAVOENZYME VANILLYL-ALCOHOL OXIDASE: Ile238Thr Mutant [Penicillium simplicissimum] |
|
| 3.85e-90 | 28 | 545 | 12 | 550 | Asp170Ser mutant of vanillyl-alcohol oxidase [Penicillium simplicissimum],1DZN_B Asp170Ser mutant of vanillyl-alcohol oxidase [Penicillium simplicissimum] |
|
| 7.59e-90 | 28 | 545 | 12 | 550 | Structure of the D170S/T457E double mutant of vanillyl-alcohol oxidase [Penicillium simplicissimum],1E0Y_B Structure of the D170S/T457E double mutant of vanillyl-alcohol oxidase [Penicillium simplicissimum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.49e-89 | 28 | 545 | 12 | 550 | Vanillyl-alcohol oxidase OS=Penicillium simplicissimum OX=69488 GN=VAOA PE=1 SV=1 |
|
| 1.68e-77 | 28 | 543 | 9 | 515 | 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit OS=Pseudomonas putida OX=303 GN=pchF PE=1 SV=3 |
|
| 3.16e-11 | 77 | 258 | 43 | 217 | Glycolate oxidase subunit GlcD OS=Bacillus subtilis (strain 168) OX=224308 GN=glcD PE=3 SV=1 |
|
| 4.06e-11 | 35 | 214 | 6 | 178 | Uncharacterized FAD-linked oxidoreductase MT2338 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2338 PE=3 SV=1 |
|
| 4.06e-11 | 35 | 214 | 6 | 178 | Uncharacterized FAD-linked oxidoreductase Rv2280 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv2280 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
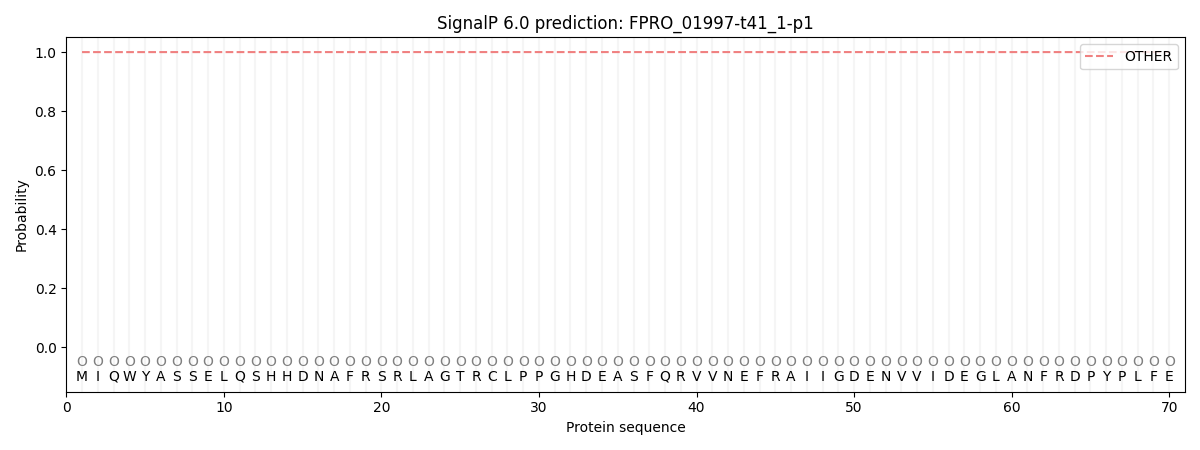
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000003 |
