You are browsing environment: FUNGIDB
CAZyme Information: FPRO_00694-t41_1-p1
You are here: Home > Sequence: FPRO_00694-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRO_00694-t41_1-p1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | related to beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 7.71e-54 | 348 | 607 | 47 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 223021 | PHA03247 | 5.93e-05 | 57 | 317 | 2702 | 2941 | large tegument protein UL36; Provisional |
| 275319 | predic_Ig_block | 9.73e-05 | 249 | 309 | 93 | 153 | putative immunoglobulin-blocking virulence protein. Members of this family are putative virulence proteins of Mycoplasma and Ureaplasma species. Members share a region of sequence similarity (see TIGR04524) with protein M, a Mycoplasma genitalium protein that binds a conserved light chain region of IgG and blocks its protective function of antigen-specific binding. The seed alignment for this model includes an N-terminal signal-anchor domain and a proline-rich linker domain, and a C-terminal extension, in addition to the protein M-like domain recognized by TIGR04524. [Cellular processes, Pathogenesis] |
| 223880 | TonB | 1.06e-04 | 251 | 325 | 44 | 118 | Periplasmic protein TonB, links inner and outer membranes [Cell wall/membrane/envelope biogenesis]. |
| 411474 | fibronec_FbpA | 3.24e-04 | 168 | 324 | 126 | 279 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.40e-97 | 146 | 607 | 147 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 7.40e-97 | 146 | 607 | 147 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 8.24e-91 | 146 | 607 | 147 | 551 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 2.04e-89 | 1 | 607 | 1 | 558 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 2.44e-88 | 23 | 607 | 32 | 601 | Probable beta-glucosidase btgE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
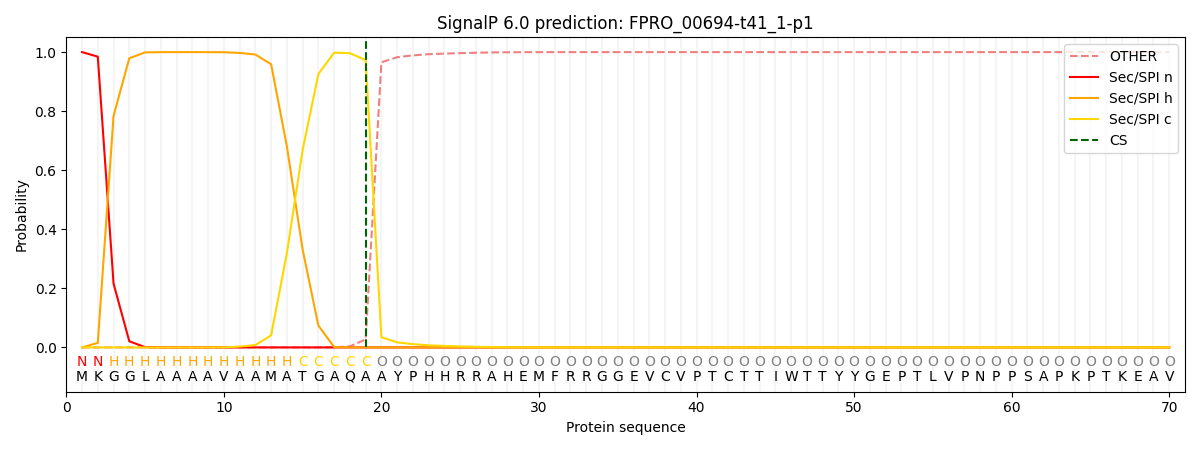
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000502 | 0.999470 | CS pos: 19-20. Pr: 0.9727 |
