You are browsing environment: FUNGIDB
CAZyme Information: FPRN_08421-t42_1-p1
You are here: Home > Sequence: FPRN_08421-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRN_08421-t42_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | related to spore coat protein SP96 precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 21 | 227 | 1.7e-63 | 0.9476439790575916 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 236090 | PRK07764 | 6.90e-12 | 255 | 356 | 404 | 505 | DNA polymerase III subunits gamma and tau; Validated |
| 237865 | PRK14951 | 1.45e-11 | 236 | 353 | 366 | 484 | DNA polymerase III subunits gamma and tau; Provisional |
| 236090 | PRK07764 | 2.30e-11 | 234 | 353 | 392 | 506 | DNA polymerase III subunits gamma and tau; Validated |
| 236090 | PRK07764 | 1.63e-10 | 263 | 353 | 389 | 480 | DNA polymerase III subunits gamma and tau; Validated |
| 237865 | PRK14951 | 4.38e-10 | 250 | 368 | 364 | 469 | DNA polymerase III subunits gamma and tau; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.44e-244 | 1 | 405 | 1 | 409 | |
| 8.05e-238 | 1 | 405 | 1 | 360 | |
| 1.06e-236 | 1 | 405 | 1 | 401 | |
| 1.06e-236 | 1 | 405 | 1 | 401 | |
| 1.06e-236 | 1 | 405 | 1 | 401 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-35 | 21 | 227 | 1 | 198 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
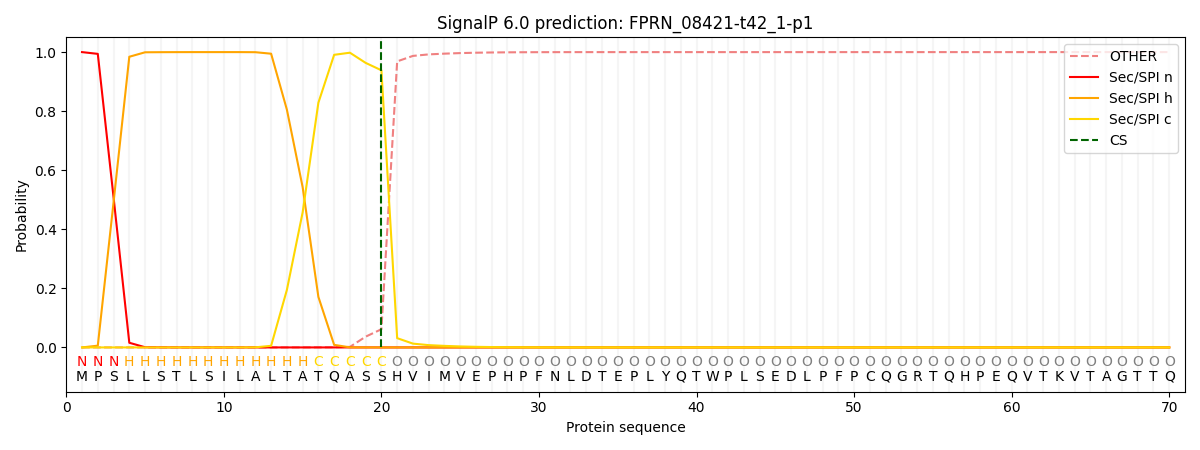
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000240 | 0.999737 | CS pos: 20-21. Pr: 0.9384 |
