You are browsing environment: FUNGIDB
FPRN_06502-t42_1-p1
Basic Information
help
Species
Fusarium proliferatum
Lineage
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum
CAZyme ID
FPRN_06502-t42_1-p1
CAZy Family
GH105
CAZyme Description
uncharacterized protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
778
89779.42
6.6123
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_FproliferatumNRRL62905
15602
N/A
348
15254
Gene Location
No EC number prediction in FPRN_06502-t42_1-p1.
Family
Start
End
Evalue
family coverage
GH139
10
775
8.2e-248
0.9781209781209781
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
408731
DUF5703_N
5.79e-141
29
312
1
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
778
1
778
0.0
1
778
1
778
0.0
1
778
1
778
0.0
1
778
1
778
0.0
5
778
5
777
FPRN_06502-t42_1-p1 has no PDB hit.
FPRN_06502-t42_1-p1 has no Swissprot hit.
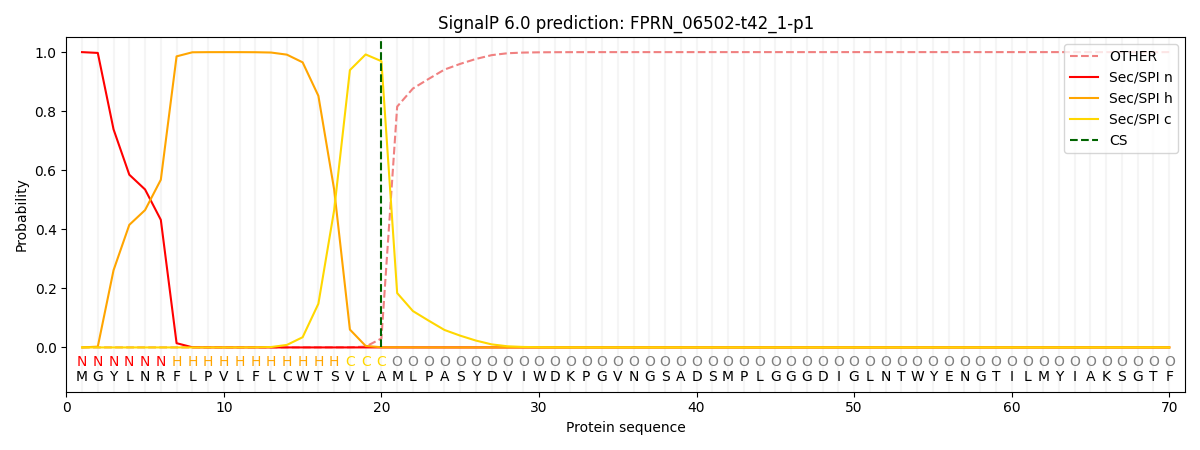
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.001062
0.998917
CS pos: 20-21. Pr: 0.9693