You are browsing environment: FUNGIDB
CAZyme Information: FPRN_00112-t42_1-p1
You are here: Home > Sequence: FPRN_00112-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRN_00112-t42_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 518960; End:524435 Strand: - | |||||||||||
Full Sequence Download help
| MNTGNAPGNI AFSSAVHLAS SISWSLHQLS TIPALPLPTN ITAVPEYEIV SKLAPALPPI | 60 |
| QVGASFGDVD DMIAGMIAAT NVSSPDKSIV GRQSTLRVMV VGDSMSQGRE GDFTWRYRIW | 120 |
| QFFKQNSIDV EMVGPYKGTV PPDPPTAPSP PILYGKPVPT SAPKTSGGYA ADVDEAFLAN | 180 |
| CNHFSAWGMS YNNIRVVFSQ CRAVAVDKGL IQTVLQATPA DLILFMLGFN DMGWFYSDAA | 240 |
| GTLDSVDTFV ANARAANPNV KIAMADVPQR SFIRGRDDLV KNTEIYNKLL PGYISKWSTK | 300 |
| QSPIHLVQLA DNYVGILIQL HTRTDGLHPN AWGEFQIAAA FSKTLVEDFN LGTSPVEIPS | 360 |
| QDDSSLGRPL QGPSNFRVFS SPQGVTATWD PVYGAYSYDV TVSINGGANT YSPTTTSSNR | 420 |
| WDSQWPLEGW TYTVSVRARY GDYSVGPYTS KLSAKATPKL SDPPQNVKVA PAADGFTVTW | 480 |
| DPPTGPNTDS IVEYNILYWD WNPDDCEIIT GAAFTSSPAA IKGLTPGRNY LIAPVTWNEN | 540 |
| GQGLPFFANN AVPGAGTPPV PSDLKILSAD STSVHITWTG SDSAGGYHVW NRNVNQKDSK | 600 |
| FEIVSNVTGS TCLDDFLLFP GTWNYEWCIS AYNGNEESPQ GPAKLAPSPA SGVDKDASAP | 660 |
| TCPTAAPWCP AGASVSVPPG QGGTLPPTTG VETITTNGMT LTITTLPAST DGDVTSTMTT | 720 |
| VPPGVSLEDF TDFPVSTNIW ITTTGKDSST TVVPVILPCP SCEPVIVWDT PEIPKVKFKW | 780 |
| PKFPELPTFH LPCIKVFGIK VSGQCPKPEG PAPINDGPPP RPRPSGSPPS DPGDDPCDFD | 840 |
| MKSGMCDNGN YPVYDPGSNT ISCDVPSDQL GSKVSQCQSH IDSKLDAVKD YLQGERSCCP | 900 |
| ASSKAKRQES LGLFSSLINR SSGSFGFRLG KRAGDYCPAK NEDPRNPPTG KCYATYTCPD | 960 |
| KLYPNVCGNA KSAINIRGAT SILTHQTGSK LHDTSPWYKG RWKATSGRPQ GGWKLDKCQV | 1020 |
| EEYPFGSGNP DRKASGNSVL RLIPGGGWED GLENQLQGAS LSRWIRTLVK RYNEFQKELT | 1080 |
| GTDPKVDGHG FVYCVEFDPG FTNYLPEDLA DQNFCASPYG VEFTLVNDYK VRGVRTWDPW | 1140 |
| FDVPGTDRKK PELIEYNDPN GDVVVTNPSG DDYAVYAVLP PTYCRYPSPG QQKWDKTTQS | 1200 |
| WSLDPRFTSY GRRDDIASFQ RCTDPPADAD DDDSDSDRLS KRYDNQTQGT MQAEKRTHAR | 1260 |
| DLISRQGSSG GFHDPNIYQW LGCGSPDDDI IEDADGDPCA DDSNPCGDGA TNLATTNGVA | 1320 |
| QSLDPLPASG VSTGAITSWP PHYTATDAPS YVPCSTHNAD PDGGIARGYC VCSGSTFALS | 1380 |
| TDSNATPANR CAYTSPLPKT TVSVSQLPTG PTASAPSPSP YVLVIYTREI EVAGHGGVTD | 1440 |
| TWWWDCRPFD KGDQSDSDVC AIKDNPIVYS TAIGRASPSQ YPTSLATFSL HGHSSCSYQG | 1500 |
| PNTATVGSVV CDDGLTAKCT AAPEASQTSF VDCTDGDEVP NGLARDL | 1547 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 96 | 345 | 7e-42 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 1.75e-31 | 180 | 345 | 14 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 238020 | FN3 | 8.26e-09 | 462 | 545 | 2 | 85 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| 214495 | FN3 | 1.30e-07 | 462 | 543 | 2 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| 394996 | fn3 | 2.17e-05 | 462 | 545 | 1 | 84 | Fibronectin type III domain. |
| 214495 | FN3 | 2.44e-05 | 558 | 638 | 1 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UKZ09962.1|CE0 | 1.94e-123 | 93 | 651 | 509 | 1055 |
| SCG40251.1|CE0 | 5.35e-98 | 88 | 638 | 328 | 856 |
| CAK39824.1|CE0 | 4.18e-83 | 14 | 376 | 59 | 395 |
| BAE63679.1|CE0 | 4.76e-83 | 93 | 375 | 54 | 317 |
| BAE60780.1|CE0 | 1.60e-78 | 148 | 391 | 19 | 244 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5B5L_A | 7.31e-09 | 165 | 347 | 26 | 201 | Crystal structure of acetyl esterase mutant S10A with acetate ion [Talaromyces cellulolyticus] |
| 5B5S_A | 7.31e-09 | 165 | 347 | 26 | 201 | Crystal structure of a carbohydrate esterase family 3 from Talaromyces cellulolyticus [Talaromyces cellulolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q63870|CO7A1_MOUSE | 1.74e-06 | 325 | 589 | 473 | 718 | Collagen alpha-1(VII) chain OS=Mus musculus OX=10090 GN=Col7a1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
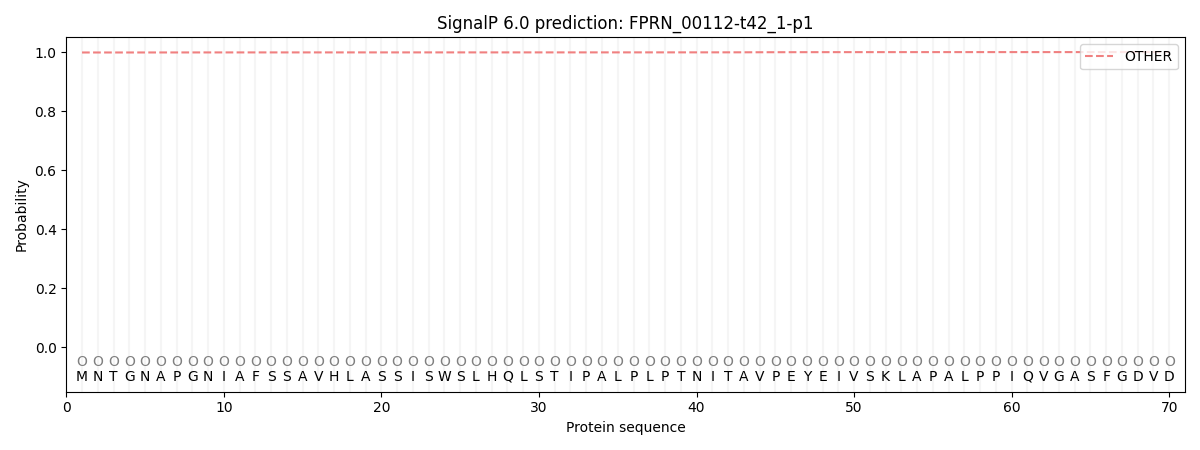
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998811 | 0.001199 |
