You are browsing environment: FUNGIDB
CAZyme Information: FPRN_00105-t42_1-p1
You are here: Home > Sequence: FPRN_00105-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRN_00105-t42_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | related to xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 542097; End:544455 Strand: + | |||||||||||
Full Sequence Download help
| MLLNLQVAAS VLSLSLLNGR AIAATPFYTL PDCTKEPLSK NGICDTSLSP PQRAAALVAA | 60 |
| LTPQEKVGNL VSNATGAPRI GLPRYNWWNE ALHGVAGSPG AHFDSNTPPY DVATSFPMPL | 120 |
| LLAAAFDDDL IHDIGNTVGT EARAFTNGGM RGVDFWTPNV NPFKDPRWGR GSETPGEDAL | 180 |
| HVSRYARNIV RGLEGDKEQR RIVATCKHYA GNDFEDWGGF TRHDFDAKIT PQDLAEYYVR | 240 |
| PFQECTRDAK VGSIMCAYNA VNGIPSCANS YLQETILRGH WNWTRDNNWI TSDCGAMQDI | 300 |
| WQNHKYVKTN AEGAQVAFEN GMDSSCEYTT TSDVSDSYKQ GLLTEKLMDR SLKRLFEGLV | 360 |
| HTGFFDGAKA QWNSLSFADV NTKKAQDLAL RSAVEGAVLL KNDGTLPLKL KKKDSVAMIG | 420 |
| FWANDTSKLQ GGYSGRAPFL HSPLYAAEKL GLDTNVAWGP TLQNSSSHDN WTTNAVAAAK | 480 |
| KSDYILYFGG LDASAAGEDR DRENLDWPES QLTLLQKLSS LGKPLVVIQL GDQVDDTALL | 540 |
| KNKKINSILW VNYPGQDGGT AVMDLLTGRK SPAGRLPVTQ YPSKYTEQIG MTDMDLRPTK | 600 |
| SLPGRTYRWY STPVLPYGFG LHYTSFKANF KANKLTFDIQ KLLKGCSAQY SDTCALPTIQ | 660 |
| VSVKNTGRIT SDFVSLVFIK SEVGPKPYPL KTLAAYGRLH DVAPSSTKDI SLEWTLDNIA | 720 |
| RRGENGDLVV YPGTYTLLLD EPTQAKVQVT LTGKKATLDK WPQDPKSA | 768 |
Enzyme Prediction help
| EC | 3.2.1.37:8 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 76 | 326 | 1.8e-60 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178629 | PLN03080 | 1.86e-171 | 11 | 739 | 14 | 768 | Probable beta-xylosidase; Provisional |
| 185053 | PRK15098 | 1.28e-55 | 50 | 735 | 30 | 746 | beta-glucosidase BglX. |
| 396478 | Glyco_hydro_3_C | 2.79e-49 | 397 | 623 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| 224389 | BglX | 7.14e-48 | 111 | 434 | 78 | 372 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| 395747 | Glyco_hydro_3 | 4.48e-26 | 79 | 355 | 63 | 313 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCT61496.1|GH3 | 0.0 | 1 | 768 | 1 | 766 |
| QGI75147.1|GH3 | 0.0 | 1 | 768 | 1 | 766 |
| QKD48431.1|GH3 | 0.0 | 1 | 768 | 1 | 766 |
| QGI88840.1|GH3 | 0.0 | 1 | 768 | 1 | 766 |
| QGI57929.1|GH3 | 0.0 | 1 | 768 | 1 | 766 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC6_A | 8.64e-248 | 31 | 763 | 3 | 741 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC7_A | 3.48e-247 | 31 | 763 | 3 | 741 | Chain A, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 5A7M_A | 2.50e-207 | 24 | 762 | 23 | 764 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 5AE6_A | 2.58e-207 | 24 | 762 | 23 | 764 | Chain A, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 6Q7I_A | 1.30e-206 | 29 | 767 | 28 | 767 | GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q0CB82|BXLB_ASPTN | 1.18e-265 | 4 | 768 | 5 | 762 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bxlB PE=3 SV=1 |
| sp|B0Y0I4|BXLB_ASPFC | 8.26e-265 | 13 | 767 | 12 | 770 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bxlB PE=3 SV=1 |
| sp|Q4WFI6|BXLB_ASPFU | 8.26e-265 | 13 | 767 | 12 | 770 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bxlB PE=3 SV=1 |
| sp|A1DJS5|XYND_NEOFI | 3.08e-262 | 29 | 767 | 28 | 770 | Probable exo-1,4-beta-xylosidase xlnD OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnD PE=3 SV=1 |
| sp|A1CCL9|BXLB_ASPCL | 1.00e-260 | 13 | 765 | 12 | 768 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=bxlB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
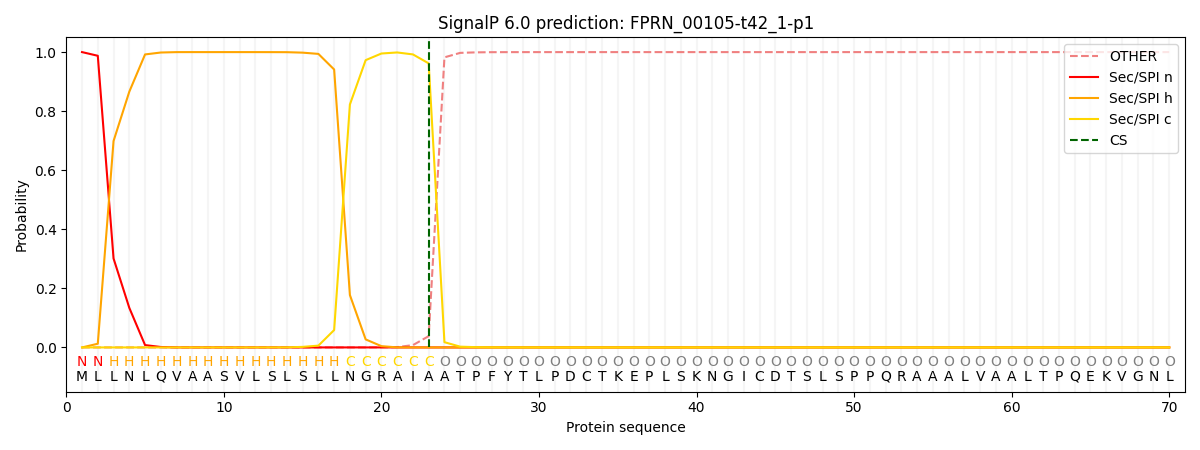
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000456 | 0.999532 | CS pos: 23-24. Pr: 0.9618 |
