You are browsing environment: FUNGIDB
CAZyme Information: FPRN_00096-t42_1-p1
You are here: Home > Sequence: FPRN_00096-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
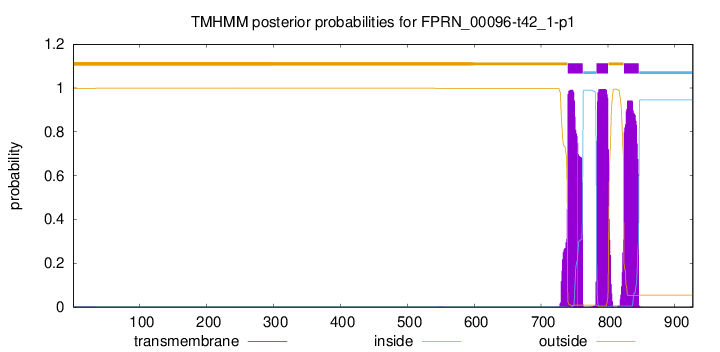
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRN_00096-t42_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | related to Melibiase subfamily | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 569514; End:572610 Strand: + | |||||||||||
Full Sequence Download help
| MADNPVFISW QTTSLTINFV KNEFGAICLY EILPLNHRHQ KSASPLFASS ELPLVSVRLN | 60 |
| GEGNTNDKTA KSLVGGYLSS RLKYKSHQQS HDADMQRLDI HSEDERAGIS VTAHLSVYRD | 120 |
| VPVLRSAVII RNESKSSDVI VTQISSLTIG GLTTRSNEWN KNYALRTATN SWFREAQWRK | 180 |
| HSLPDIGIDK NGICELHDGH SGSQATYGLQ NRGSFSTGSY LPMGILESES KSDTWAWQIE | 240 |
| HNGSWRWEIG DYKDSVYIAA GGPTKADHDW RHTLCPGDSF TSPPVSLTRV SGCFQDAIRA | 300 |
| LNDYRRRIIR PHDDNKRLPI IFNDYMNCLM GDPDEDKIEA LLDPVAQSGA EYFVIDAGWY | 360 |
| ADDGNWWDDV GLWEPSKRRF PSGFKTLMDK IKSKGLIPGL WLEPEVVGVR SVVGERLPED | 420 |
| AFFHENGQRV VERGRFQLDY RHPEVRAWMT SVIDRLVVHY GAGFFKFDYN IEVIQGTDAP | 480 |
| GPSSAGANQL LHQRCYLDWV RSLLDKHPNL VIENCSSGAQ RMDYAMLSVH SLQSTSDQQD | 540 |
| PILYAAIAAA LPTCVLPEQS ASWAYPQPEW SDELNAFTVV NSLLGRVYLS GRIDRLSPSQ | 600 |
| MELIIEGMDV YKTIRQHLVT AHAIWPLGLP RWHDEWISLG LETDNGLYLS VWRRGGSCSM | 660 |
| EIPLPRFAGS RNVQVNVLYP KRLPTQAIWN EGNDMLELKF PETRTSGDAD SLRSNHPGAM | 720 |
| SYCMNYNIKM GFTHIKFSLI FVHVLTFLLF TDWALDTLHG FYEKDCPPPK KGSLVRSLIK | 780 |
| KKSLAILTAF LALVPMAFRL ADLVRFYKGL RDRYTPEGGL PWSFGQVTAV AVWLPVVCKF | 840 |
| LHYCVVKDGV QARIDKDEYV VSRRETAISE FKSSEEPGLD NVSLIRPAVP GSKVEAEPIE | 900 |
| MRASLDASAE SLLMIPHAGW SPDMETP | 927 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 75 | 621 | 3e-97 | 0.7747093023255814 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 269892 | GH36 | 2.23e-84 | 317 | 613 | 1 | 299 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 307952 | Melibiase | 4.82e-34 | 279 | 542 | 1 | 265 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| 225882 | GalA | 3.26e-28 | 49 | 542 | 51 | 510 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| 269893 | GH27 | 3.93e-10 | 322 | 396 | 5 | 80 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 407116 | Glyco_hydro_36N | 3.94e-06 | 76 | 183 | 68 | 181 | Glycosyl hydrolase family 36 N-terminal domain. This domain is found at the N-terminus of many family 36 glycoside hydrolases. It has a beta-supersandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCT61505.1|GH36 | 0.0 | 1 | 704 | 1 | 704 |
| QGI88848.1|GH36 | 0.0 | 1 | 704 | 1 | 704 |
| QGI57937.1|GH36 | 0.0 | 1 | 704 | 1 | 704 |
| QGI75155.1|GH36 | 0.0 | 1 | 704 | 1 | 699 |
| QKD48422.1|GH36 | 0.0 | 1 | 704 | 1 | 704 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PHU_A | 3.07e-23 | 94 | 615 | 143 | 642 | SpAga wild type apo structure [Streptococcus pneumoniae TIGR4],6PHV_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4] |
| 2XN0_A | 5.25e-23 | 92 | 616 | 131 | 634 | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN0_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN1_A Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_C Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_D Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM] |
| 2XN2_A | 6.92e-23 | 92 | 616 | 131 | 634 | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose [Lactobacillus acidophilus NCFM] |
| 6PHW_A | 1.23e-22 | 94 | 615 | 143 | 642 | Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PHX_A SpAga D472N structure in complex with raffinose [Streptococcus pneumoniae TIGR4],6PHY_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PI0_A AgaD472N-Linear Blood group B type 2 trisaccharide complex structure [Streptococcus pneumoniae TIGR4] |
| 4FNQ_A | 2.08e-22 | 63 | 647 | 80 | 662 | Crystal structure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q5ARP5|AGALG_EMENI | 1.26e-23 | 183 | 542 | 189 | 552 | Probable alpha-galactosidase G OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglG PE=2 SV=1 |
| sp|P43469|AGAL2_PEDPE | 1.50e-22 | 269 | 665 | 279 | 672 | Alpha-galactosidase 2 OS=Pediococcus pentosaceus OX=1255 GN=agaS PE=3 SV=1 |
| sp|G1UB44|MELA_LACAC | 2.70e-22 | 92 | 616 | 131 | 634 | Alpha-galactosidase Mel36A OS=Lactobacillus acidophilus (strain ATCC 700396 / NCK56 / N2 / NCFM) OX=272621 GN=melA PE=1 SV=1 |
| sp|P27756|AGAL_STRMU | 7.97e-22 | 68 | 566 | 101 | 560 | Alpha-galactosidase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=aga PE=3 SV=3 |
| sp|Q9ALJ4|AGAA_GEOSE | 1.87e-21 | 63 | 647 | 80 | 662 | Alpha-galactosidase AgaA OS=Geobacillus stearothermophilus OX=1422 GN=agaA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
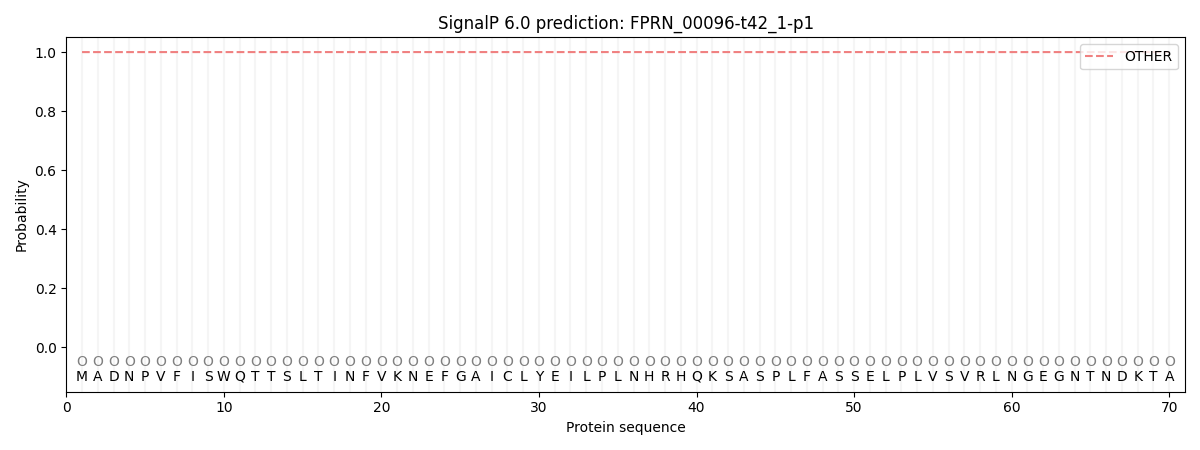
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000064 | 0.000000 |