You are browsing environment: FUNGIDB
CAZyme Information: FPRN_00008-t42_1-p1
You are here: Home > Sequence: FPRN_00008-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium proliferatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium proliferatum | |||||||||||
| CAZyme ID | FPRN_00008-t42_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | probable Dextranase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 782705; End:784492 Strand: + | |||||||||||
Full Sequence Download help
| MRGLAVFAGL FISLVGAAPS NHNAYDNRAH CSSDFCTWWH SEGEINTDSP VKPENVRQSH | 60 |
| QYTVQVSAAG ANKFYDSFVY EAIPRNGNGR IYAPTDAPNS NTLGTDVDDG ITIEPKIGLN | 120 |
| MAWSQFEYSQ DVDVKIKRRD GTALRANDVI IRPVSISYEL SQSRDGGLIV RVPKDNNGRR | 180 |
| MSVEFNSDLY TFRSDGNQYV TSGGSVVGRE PRNALAIFAS PPLPSNKKPR MSSSNTQTMK | 240 |
| PGPINNGDWG TKPILYFPPG VYYMNQDQKG KSGKLGSNHI RLSPNTYWVY FAPGAYVKGA | 300 |
| IEYSTKQDFY ATGLGVLSGE HYVYQANAAK GYVAEKSDQY GLRMWWHNSI NQGQKWTCQG | 360 |
| PTVAAPPFNT MDFNGNSDIT SEIVDYKQVG AYFFQTDGPA IYPNSVVHDI FYHVNDDGIK | 420 |
| MYYSGVSVSR ATVWKCHNDP IIQMGWDTRN LDNIKIDTLN VIHTRYYKSE TVVPSAIIGA | 480 |
| SPFYMDGKSA DPSKSISATI SNIVCEGPCP GLLRITPLQS YKNFVIKNVA FPDGLQTNVI | 540 |
| GIGQSIVPKQ SGVTMDLEID NWTVSGQKVT MQNFQSDKLG QLNIDGSYWG QWKIT | 595 |
Enzyme Prediction help
| EC | 3.2.1.11:4 | 3.2.1.57:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH49 | 29 | 593 | 1.3e-253 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 407501 | Glyco_hydro_49N | 6.76e-80 | 30 | 221 | 4 | 185 | Glycosyl hydrolase family 49 N-terminal Ig-like domain. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the N-terminal Ig-like fold. |
| 397674 | Glyco_hydro_49 | 8.68e-33 | 487 | 594 | 3 | 117 | Glycosyl hydrolase family 49. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the C-terminal pectate lyase like domain. |
| 408611 | B_solenoid_dext | 4.46e-13 | 367 | 398 | 1 | 33 | Beta solenoid repeat from Dextranase. The crystal structures of Dex49A from Penicillium minioluteum and of ATCC9642 isopullulanase from Aspergillus niger include beta-solenoid repeats, sharing structural similarities. This Pfam entry includes a single repeat unit of the repeat regions. |
| 408554 | IPU_b_solenoid | 6.27e-10 | 434 | 466 | 1 | 33 | Isopullulanase beta-solenoid repeat. IPU and dextranase repeat unit includes three (or one long and one short) parallel beta-strands. The repeat region as a whole folds into a beta-helix, known as beta-solenoid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA21309.1|GH49 | 0.0 | 1 | 595 | 2 | 606 |
| AAB47720.1|GH49|3.2.1.11 | 0.0 | 1 | 595 | 5 | 608 |
| CAB91097.1|GH49 | 0.0 | 1 | 595 | 5 | 608 |
| AHJ14526.1|GH49 | 0.0 | 19 | 595 | 31 | 608 |
| AZC11123.1|GH49 | 0.0 | 19 | 595 | 31 | 608 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1OGM_X | 0.0 | 28 | 595 | 6 | 574 | Dex49A from Penicillium minioluteum [Talaromyces minioluteus],1OGO_X Dex49A from Penicillium minioluteum complex with isomaltose [Talaromyces minioluteus] |
| 1WMR_A | 9.79e-108 | 32 | 590 | 10 | 543 | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1WMR_B Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_A Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_B Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],2Z8G_A Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger],2Z8G_B Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger] |
| 3WWG_A | 2.74e-107 | 32 | 590 | 10 | 543 | Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_B Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_C Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_D Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger] |
| 6NZS_A | 1.09e-94 | 37 | 589 | 12 | 579 | Dextranase AoDex KQ11 [Pseudarthrobacter oxydans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P48845|DEXT_TALMI | 0.0 | 1 | 595 | 5 | 608 | Dextranase OS=Talaromyces minioluteus OX=28574 GN=DEX PE=1 SV=1 |
| sp|O00105|IPUA_ASPNG | 7.73e-107 | 32 | 590 | 25 | 558 | Isopullulanase OS=Aspergillus niger OX=5061 GN=ipuA PE=1 SV=1 |
| sp|P70744|DEXT_ARTGO | 5.22e-95 | 16 | 592 | 37 | 633 | Dextranase OS=Arthrobacter globiformis OX=1665 PE=3 SV=1 |
| sp|P39652|DEXT_ARTSD | 3.24e-92 | 37 | 589 | 63 | 630 | Dextranase OS=Arthrobacter sp. (strain CB-8) OX=74565 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
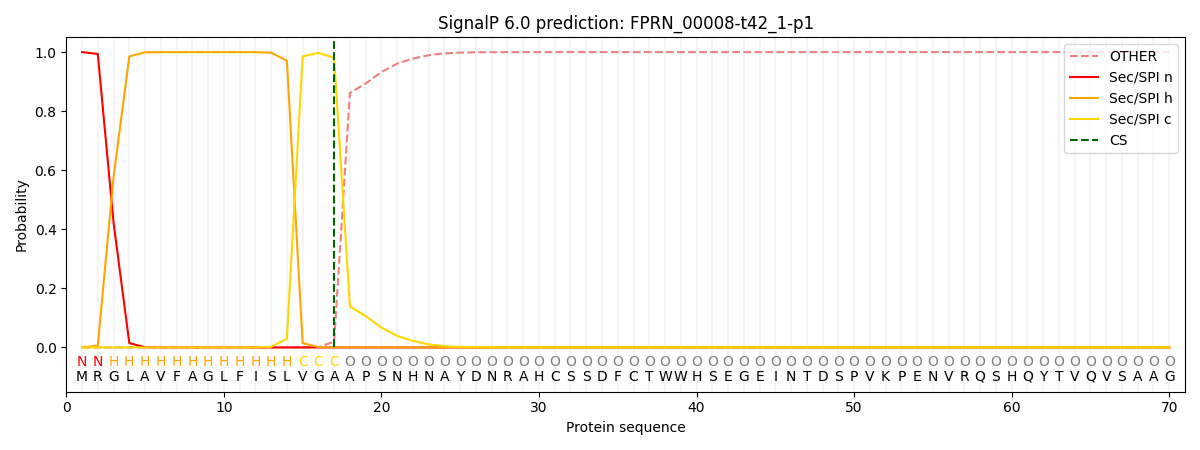
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000218 | 0.999758 | CS pos: 17-18. Pr: 0.9805 |
