You are browsing environment: FUNGIDB
CAZyme Information: FOZG_16353-t36_1-p1
You are here: Home > Sequence: FOZG_16353-t36_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
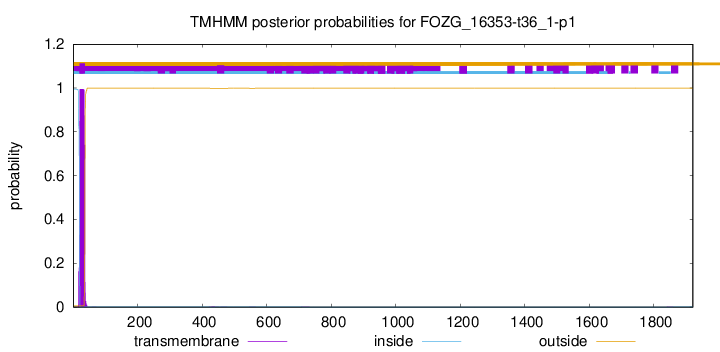
TMHMM annotations
Basic Information help
| Species | Fusarium oxysporum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium oxysporum | |||||||||||
| CAZyme ID | FOZG_16353-t36_1-p1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 293786 | ANK | 1.25e-25 | 1093 | 1181 | 1 | 89 | ankyrin repeats. Ankyrin repeats are one of the most abundant repeat motifs, and generally function as scaffolds for protein-protein interactions in processes including cell cycle, transcriptional regulation, signal transduction, vesicular trafficking, and inflammatory response. Although predominantly found in eukaryotic proteins, they are also found in some bacterial and viral proteins. Less is known of their physiological roles in prokaryotes. Some bacterial ANK proteins play key roles in microbial pathogenesis by mimicking or manipulating host function(s). The pathogen Providencia alcalifaciens N-formyltransferase ankyrin repeats function in small molecule binding and allosteric control. Ankyrin-repeat proteins have been associated with a number of human diseases. |
| 222980 | PHA03095 | 1.90e-19 | 1344 | 1694 | 27 | 316 | ankyrin-like protein; Provisional |
| 293786 | ANK | 4.70e-18 | 1055 | 1146 | 5 | 87 | ankyrin repeats. Ankyrin repeats are one of the most abundant repeat motifs, and generally function as scaffolds for protein-protein interactions in processes including cell cycle, transcriptional regulation, signal transduction, vesicular trafficking, and inflammatory response. Although predominantly found in eukaryotic proteins, they are also found in some bacterial and viral proteins. Less is known of their physiological roles in prokaryotes. Some bacterial ANK proteins play key roles in microbial pathogenesis by mimicking or manipulating host function(s). The pathogen Providencia alcalifaciens N-formyltransferase ankyrin repeats function in small molecule binding and allosteric control. Ankyrin-repeat proteins have been associated with a number of human diseases. |
| 293786 | ANK | 1.98e-17 | 1330 | 1433 | 1 | 98 | ankyrin repeats. Ankyrin repeats are one of the most abundant repeat motifs, and generally function as scaffolds for protein-protein interactions in processes including cell cycle, transcriptional regulation, signal transduction, vesicular trafficking, and inflammatory response. Although predominantly found in eukaryotic proteins, they are also found in some bacterial and viral proteins. Less is known of their physiological roles in prokaryotes. Some bacterial ANK proteins play key roles in microbial pathogenesis by mimicking or manipulating host function(s). The pathogen Providencia alcalifaciens N-formyltransferase ankyrin repeats function in small molecule binding and allosteric control. Ankyrin-repeat proteins have been associated with a number of human diseases. |
| 403870 | Ank_2 | 2.24e-16 | 1098 | 1181 | 1 | 82 | Ankyrin repeats (3 copies). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 160 | 1399 | 1 | 1207 | |
| 0.0 | 160 | 1399 | 1 | 1069 | |
| 0.0 | 160 | 1399 | 1 | 1069 | |
| 4.17e-252 | 1 | 409 | 1 | 407 | |
| 9.40e-212 | 1 | 406 | 1 | 406 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.45e-12 | 1330 | 1812 | 63 | 540 | Transient receptor potential cation channel subfamily A member 1 OS=Rattus norvegicus OX=10116 GN=Trpa1 PE=2 SV=1 |
|
| 1.14e-10 | 1330 | 1812 | 63 | 540 | Transient receptor potential cation channel subfamily A member 1 OS=Mus musculus OX=10090 GN=Trpa1 PE=1 SV=1 |
|
| 3.57e-10 | 600 | 943 | 273 | 622 | Vegetative incompatibility protein HET-E-1 OS=Podospora anserina OX=2587412 GN=HET-E1 PE=4 SV=1 |
|
| 3.00e-09 | 110 | 234 | 57 | 178 | Inositol phosphoceramide mannosyltransferase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC17G8.11c PE=1 SV=1 |
|
| 5.07e-07 | 1330 | 1475 | 136 | 280 | Ankyrin repeat domain-containing protein 16 OS=Mus musculus OX=10090 GN=Ankrd16 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.840495 | 0.159511 |