You are browsing environment: FUNGIDB
CAZyme Information: FOYG_01057-t42_2-p1
Basic Information
help
| Species |
Fusarium oxysporum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium oxysporum
|
| CAZyme ID |
FOYG_01057-t42_2-p1
|
| CAZy Family |
AA3 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 328 |
|
36163.62 |
4.6007 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FoxysporumNRRL32931 |
17642 |
660029 |
362 |
17280
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH12 |
181 |
324 |
3.6e-24 |
0.8717948717948718 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396303
|
Glyco_hydro_12 |
3.93e-16 |
75 |
325 |
3 |
207 |
Glycosyl hydrolase family 12. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.32e-247 |
1 |
328 |
1 |
328 |
| 3.21e-239 |
1 |
328 |
1 |
328 |
| 3.21e-239 |
1 |
328 |
1 |
328 |
| 6.47e-239 |
1 |
328 |
1 |
328 |
| 7.55e-238 |
1 |
328 |
1 |
328 |
FOYG_01057-t42_2-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.15e-13 |
80 |
324 |
39 |
240 |
Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=xgeA PE=3 SV=1 |
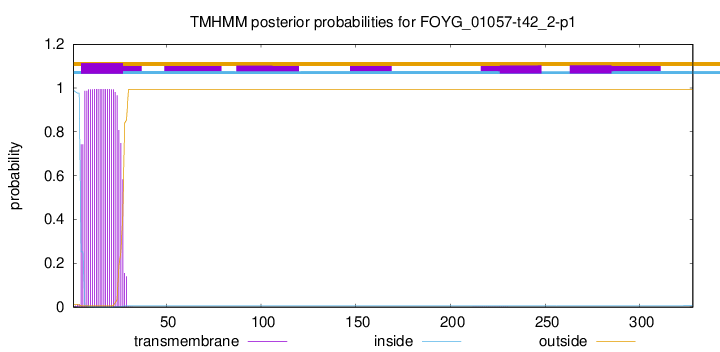
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 0.899737 |
0.100267 |
|