You are browsing environment: FUNGIDB
CAZyme Information: FOC4_g10006766-t38_1-p1
You are here: Home > Sequence: FOC4_g10006766-t38_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
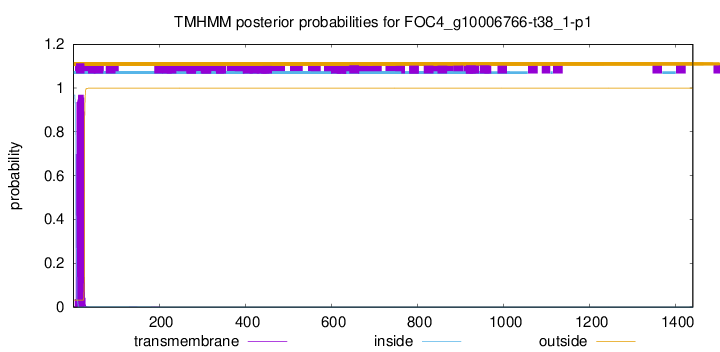
TMHMM annotations
Basic Information help
| Species | Fusarium odoratissimum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium odoratissimum | |||||||||||
| CAZyme ID | FOC4_g10006766-t38_1-p1 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | AP-1 complex subunit gamma-1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 104 | 606 | 7.3e-164 | 0.9932735426008968 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 0.0 | 104 | 607 | 2 | 453 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 396262 | Adaptin_N | 5.52e-151 | 651 | 1193 | 1 | 520 | Adaptin N terminal region. This family consists of the N terminal region of various alpha, beta and gamma subunits of the AP-1, AP-2 and AP-3 adaptor protein complexes. The adaptor protein (AP) complexes are involved in the formation of clathrin-coated pits and vesicles. The N-terminal region of the various adaptor proteins (APs) is constant by comparison to the C-terminal which is variable within members of the AP-2 family; and it has been proposed that this constant region interacts with another uniform component of the coated vesicles. |
| 240427 | PTZ00470 | 5.73e-122 | 91 | 611 | 67 | 522 | glycoside hydrolase family 47 protein; Provisional |
| 197886 | Alpha_adaptinC2 | 1.06e-24 | 1336 | 1437 | 2 | 102 | Adaptin C-terminal domain. Adaptins are components of the adaptor complexes which link clathrin to receptors in coated vesicles. Clathrin-associated protein complexes are believed to interact with the cytoplasmic tails of membrane proteins, leading to their selection and concentration. Gamma-adaptin is a subunit of the golgi adaptor. Alpha adaptin is a heterotetramer that regulates clathrin-bud formation. The carboxyl-terminal appendage of the alpha subunit regulates translocation of endocytic accessory proteins to the bud site. This Ig-fold domain is found in alpha, beta and gamma adaptins and consists of a beta-sandwich containing 7 strands in 2 beta-sheets in a greek-key topology.. The adaptor appendage contains an additional N-terminal strand. |
| 397151 | Alpha_adaptinC2 | 3.66e-20 | 1336 | 1435 | 6 | 108 | Adaptin C-terminal domain. Alpha adaptin is a heterotetramer which regulates clathrin-bud formation. The carboxyl-terminal appendage of the alpha subunit regulates translocation of endocytic accessory proteins to the bud site. This ig-fold domain is found in alpha, beta and gamma adaptins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1439 | 1 | 1429 | |
| 0.0 | 5 | 1438 | 4 | 1469 | |
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 | |
| 0.0 | 1 | 610 | 1 | 610 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.48e-190 | 635 | 1207 | 4 | 583 | Structure of the cargo bound AP-1:Arf1:tetherin-Nef stable closed trimer [Mus musculus],6CRI_Q Structure of the cargo bound AP-1:Arf1:tetherin-Nef stable closed trimer [Mus musculus],6CRI_R Structure of the cargo bound AP-1:Arf1:tetherin-Nef stable closed trimer [Mus musculus] |
|
| 2.55e-190 | 635 | 1207 | 7 | 586 | Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1 [Mus musculus],6CM9_G Structure of the cargo bound AP-1:Arf1:tetherin-Nef closed trimer monomeric subunit [Mus musculus],6D83_G Structure of the cargo bound AP-1:Arf1:tetherin-Nef (L164A, L165A) dileucine mutant dimer monomeric subunit [Mus musculus],6D84_G Structure of the cargo bound AP-1:Arf1:tetherin-Nef (L164A, L165A) dileucine mutant dimer [Mus musculus],6D84_K Structure of the cargo bound AP-1:Arf1:tetherin-Nef (L164A, L165A) dileucine mutant dimer [Mus musculus],6DFF_G Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer [Mus musculus] |
|
| 4.37e-190 | 623 | 1207 | 11 | 600 | Crystal structure of the human BST2 cytoplasmic domain and the HIV-1 Vpu cytoplasmic domain bound to the clathrin adaptor protein complex 1 (AP1) core [Mus musculus] |
|
| 4.53e-190 | 635 | 1207 | 12 | 591 | AP1 clathrin adaptor core [Mus musculus],1W63_C AP1 clathrin adaptor core [Mus musculus],1W63_E AP1 clathrin adaptor core [Mus musculus],1W63_G AP1 clathrin adaptor core [Mus musculus],1W63_I AP1 clathrin adaptor core [Mus musculus],1W63_K AP1 clathrin adaptor core [Mus musculus] |
|
| 3.88e-67 | 623 | 1224 | 11 | 634 | HIV-1 Nef in complex with the CD4 cytoplasmic domain and the AP2 clathrin adaptor complex [Rattus norvegicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.11e-241 | 635 | 1440 | 30 | 873 | AP-1 complex subunit gamma-1 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=APL4 PE=3 SV=3 |
|
| 1.08e-201 | 635 | 1422 | 5 | 876 | AP-1 complex subunit gamma OS=Dictyostelium discoideum OX=44689 GN=ap1g1 PE=1 SV=1 |
|
| 3.63e-195 | 635 | 1435 | 7 | 814 | AP-1 complex subunit gamma-1 OS=Homo sapiens OX=9606 GN=AP1G1 PE=1 SV=5 |
|
| 5.47e-194 | 635 | 1435 | 7 | 814 | AP-1 complex subunit gamma-1 OS=Mus musculus OX=10090 GN=Ap1g1 PE=1 SV=3 |
|
| 4.17e-193 | 635 | 1435 | 7 | 814 | AP-1 complex subunit gamma-1 OS=Pongo abelii OX=9601 GN=AP1G1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999984 | 0.000037 |