You are browsing environment: FUNGIDB
CAZyme Information: FOC1_g10009133-t38_1-p1
You are here: Home > Sequence: FOC1_g10009133-t38_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
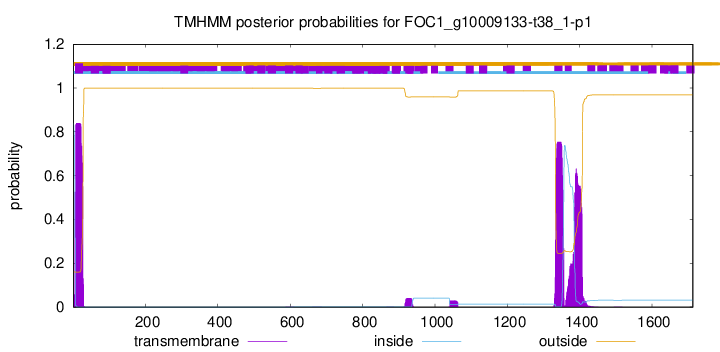
TMHMM annotations
Basic Information help
| Species | Fusarium oxysporum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium oxysporum | |||||||||||
| CAZyme ID | FOC1_g10009133-t38_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | Killer toxin subunits alpha/beta | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 270746; End:280612 Strand: + | |||||||||||
Full Sequence Download help
| MRISTFIAAV LSTVFTIYPA FASGTAAPQL NSCPPLIDEA CVDSSNWTLF TNLDEALACN | 60 |
| HKPRLLDFTI HYPLRPSGPG NVLRACTSYQ GETLNLTSVV STSSLARNNQ TKSKLELVWD | 120 |
| DTDEKVIMPQ AITALKELQS FLLTEGSKDR QSIFSQYGDT SVGLYIGDKV VPSSVANVVI | 180 |
| EALVKRLRES GMPRRLGVQL CGDGRNSDYT SGIIIDTKPG PASLINAQVA VSGWSNATCI | 240 |
| DGFQHSSNFG SFLIETIKED TAVSRPSNST APKLAKRAEC KTIQVKSGDG CGDLAQRCGI | 300 |
| KAADFTKYNS QTKNLCSTLI DGQHVCCSSG DLPDFRPKPK PDGSCATYTV ESGDFCNKIA | 360 |
| AANSLKVEDI ESFNKNTWGW SGCKLLFDKA VICLSKGDPP MPNPIANAEC GPQKPGTKKP | 420 |
| ENGKDLADLN PCPLNACCNI WGQCGVTADF CKNTTLGAPG TAKPGTNGCI SNCGTDIVNN | 480 |
| KSPPSSFIID ISHVSYQFNK FKKLKGSKRI LAFGGWVDST HPTKYHILRN AVKMENRLQV | 540 |
| AKNIAAFVKK HDLDGVDIDW EYPGAPDIPD IPKADEEDGK NYLGLLTLLK GQLGGLSLSI | 600 |
| AAPASFWYLK PYPINNIAKV VDYIIDMTYD LHGQWDYDNK WASPGCPKGN CLRSHVNLTE | 660 |
| TMGALAMITK AGVPSTKVIV GVTSYGRSFK MAKKGCVGPI DSDILVYNDT EWVAYMSDST | 720 |
| KKRRISKYQG LNFGGVTDWA VDLQKFGKLE DSNLIEIIDK DGKCKWKTKD GFSCLDKAVI | 780 |
| ESDMNPQDRW NGVRAPCAWR DMASSWVDKR SSQNINVTGK ENANKFSRHV SDLADGYEKF | 840 |
| DCASFSVGVN GCEQEDKCED TRDSGPAGYF ILNSFTSIHT TFKSLWQTVD KVENGMQSKL | 900 |
| DQLVETFAPD VDESSEFNLI ADFLGIAVGM FAAPFFNKIM KTKDSPTADI KDLIANTASW | 960 |
| GATLAKDIQN SKRETPKSKV AGKLSEISDL WTESIEAFTE KAFRGDDESV DYIGDLIADG | 1020 |
| KFNNDEVTFD MSDLRKQLRK VFFAVLIPEA WKVGAGYAPA FVMDSGYDCN AVGPLDYEYI | 1080 |
| APSTGEEMGY CYQGRRYYLL APNGRERNCD PQVPGGTGGN PTDCKPNYFT APQGIENLDP | 1140 |
| KNQDTYDWGG ITAQDLVAGA VEGWKANKEK NGGGFLDLTK TSNYDFLLDG NSDEVNIRLP | 1200 |
| GFIQIPYGGH NGNWNSTAHG GSKPSPNGGS NSGSKCGSSG GSQTSSNGGS SSGSSFGSNR | 1260 |
| GPNGSNGSGS KNTQPSIVPV ISGAAEQIWQ NTLTPFQSTH QFQTLESTFR FRQADGQELD | 1320 |
| CGRAKITPDL GDSISAVLTF VPLAILVVVT LSSWRVNQSS LTYAHGLNAS SLWSVVLDVT | 1380 |
| SYLRYLQFAF IAALMSIEYP GFFVPAVSKL SWASLLYWSG PFSNGYMYEG PTGAGHFLIQ | 1440 |
| VLDDQAAYAV WDDREASKGR FIGYLYMDLL SRDNKYKGNQ NVNLQCGYLK NDGSRVYPAT | 1500 |
| LLMCSFPPPA PSACTFLKHS QIISLFHEEL TRSELGHGIH DLLARTNYVA FHGHRSPPDF | 1560 |
| AETLSVMLEK WCWMKPELKR LGLHYTSTDL HLKEKWLREH PGEDLPPERI PDDTIERLIK | 1620 |
| GRQVTRSLWY LRQLVYAHFD MKVHHPRSHT ELRNMDSTKI FNDLLEKLWL VRAPQPEDQG | 1680 |
| YPHADLGHLV SGYDAGYYSY LRRVDLNLTI KND | 1713 |
Enzyme Prediction help
| EC | 3.2.1.14:3 | 3.2.1.14:3 | 3.2.1.14:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 490 | 742 | 8.4e-38 | 0.8243243243243243 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119357 | GH18_zymocin_alpha | 2.49e-150 | 485 | 744 | 40 | 345 | Zymocin, alpha subunit. Zymocin is a heterotrimeric enzyme that inhibits yeast cell cycle progression. The zymocin alpha subunit has a chitinase activity that is essential for holoenzyme action from the cell exterior while the gamma subunit contains the intracellular toxin responsible for G1 phase cell cycle arrest. The zymocin alpha and beta subunits are thought to act from the cell's exterior by docking to the cell wall-associated chitin, thus mediating gamma-toxin translocation. The alpha subunit has an eight-stranded TIM barrel fold similar to that of family 18 glycosyl hydrolases such as hevamine, chitolectin, and chitobiase. |
| 341050 | M3A_TOP | 4.20e-97 | 1443 | 1701 | 356 | 581 | Peptidase M3 thimet oligopeptidase (TOP), also includes neurolysin. Peptidase M3 Thimet oligopeptidase (TOP; PZ-peptidase; endo-oligopeptidase A; endopeptidase 24.15; soluble metallo-endopeptidase; EC 3.4.24.15) family also includes neurolysin (endopeptidase 24.16, microsomal endopeptidase, mitochondrial oligopeptidase M, neurotensin endopeptidase, soluble angiotensin II-binding protein, thimet oligopeptidase II) which hydrolyzes oligopeptides such as neurotensin, bradykinin and dynorphin A. TOP and neurolysin are neuropeptidases expressed abundantly in the testis, but are also found in the liver, lung and kidney. They are involved in the metabolism of neuropeptides under 20 amino acid residues long and cleave most bioactive peptides at the same sites, but recognize different positions on some naturally occurring and synthetic peptides; they cleave at distinct sites on the 13-residue bioactive peptide neurotensin, which modulates central dopaminergic and cholinergic circuits. TOP has been shown to degrade peptides released by the proteasome, limiting the extent of antigen presentation by major histocompatibility complex class I molecules, and has been associated with amyloid protein precursor processing. |
| 341068 | M3A | 1.07e-41 | 1450 | 1701 | 308 | 526 | Peptidase M3A family includes thimet oligopeptidase, dipeptidyl carboxypeptidase and mitochondrial intermediate peptidase. The M3-like family also called neurolysin-like family, is part of the "zincins" metallopeptidases, and includes M3, M2 and M32 families of metallopeptidases. The M3 family is subdivided into two subfamilies: the widespread M3A, represented by this CD, which comprises a number of high-molecular mass endo- and exopeptidases from bacteria, archaea, protozoa, fungi, plants and animals, and the small M3B, whose members are enzymes primarily from bacteria. Well-known mammalian/eukaryotic M3A endopeptidases are the thimet oligopeptidase (TOP; endopeptidase 3.4.24.15), neurolysin (alias endopeptidase 3.4.24.16), and the mitochondrial intermediate peptidase. The first two are intracellular oligopeptidases, which act only on relatively short substrates of less than 20 amino acid residues, while the latter cleaves N-terminal octapeptides from proteins during their import into the mitochondria. The M3A subfamily also contains several bacterial endopeptidases, called oligopeptidases A, as well as a large number of bacterial carboxypeptidases, called dipeptidyl peptidases (Dcp; Dcp II; peptidyl dipeptidase; EC 3.4.15.5). The peptidases in the M3 family contain the HEXXH motif that forms part of the active site in conjunction with a C-terminally-located Glutamic acid (Glu) residue. A single zinc ion is ligated by the side-chains of the two Histidine (His) residues, and the more C-terminal Glu. Most of the peptidases are synthesized without signal peptides or propeptides, and function intracellularly. |
| 396149 | Peptidase_M3 | 4.98e-41 | 1444 | 1701 | 168 | 387 | Peptidase family M3. This is the Thimet oligopeptidase family, large family of mammalian and bacterial oligopeptidases that cleave medium sized peptides. The group also contains mitochondrial intermediate peptidase which is encoded by nuclear DNA but functions within the mitochondria to remove the leader sequence. |
| 223416 | Dcp | 1.76e-40 | 1448 | 1701 | 398 | 618 | Zn-dependent oligopeptidase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKD52296.1|CBM18|GH18 | 0.0 | 147 | 1713 | 256 | 2367 |
| AEO59465.1|CBM18|CBM50|GH18 | 4.00e-257 | 32 | 1206 | 43 | 1322 |
| EGX53225.1|CBM18|CBM50|GH18 | 2.25e-255 | 24 | 1226 | 28 | 1343 |
| QMW48046.1|CBM18|CBM50|GH18 | 2.29e-255 | 33 | 1206 | 35 | 1312 |
| QRD93063.1|CBM18|CBM50|GH18 | 2.29e-255 | 33 | 1206 | 35 | 1312 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2O36_A | 3.63e-26 | 1454 | 1701 | 385 | 598 | Chain A, Thimet oligopeptidase [Homo sapiens] |
| 1S4B_P | 4.79e-26 | 1454 | 1701 | 385 | 598 | Chain P, Thimet oligopeptidase [Homo sapiens] |
| 1HKK_A | 8.13e-20 | 497 | 746 | 58 | 344 | High resoultion crystal structure of human chitinase in complex with allosamidin [Homo sapiens] |
| 1HKI_A | 8.24e-20 | 497 | 746 | 58 | 344 | Crystal structure of human chitinase in complex with glucoallosamidin B [Homo sapiens],1HKJ_A Crystal structure of human chitinase in complex with methylallosamidin [Homo sapiens],1HKM_A High resolution crystal structure of human chitinase in complex with demethylallosamidin [Homo sapiens] |
| 1LG1_A | 1.11e-19 | 497 | 746 | 58 | 344 | Crystal Structure Of Human Chitotriosidase In Complex With Chitobiose [Homo sapiens],1LG2_A Crystal Structure Of Human Chitotriosidase In Complex With Ethylene Glycol [Homo sapiens],1LQ0_A Crystal Structure Of Human Chitotriosidase At 2.2 Angstrom Resolution [Homo sapiens],6ZE8_A Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens],6ZE8_B Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens],6ZE8_C Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens],6ZE8_D Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens],6ZE8_E Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens],6ZE8_F Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound OATD-01 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P09805|KTXA_KLULA | 1.59e-87 | 120 | 696 | 50 | 623 | Killer toxin subunits alpha/beta OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 PE=1 SV=1 |
| sp|P25375|PRTD_YEAST | 1.22e-35 | 1443 | 1701 | 417 | 647 | Saccharolysin OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PRD1 PE=1 SV=1 |
| sp|Q1JPJ8|THOP1_BOVIN | 7.79e-29 | 1443 | 1701 | 392 | 613 | Thimet oligopeptidase OS=Bos taurus OX=9913 GN=THOP1 PE=2 SV=3 |
| sp|Q54DD2|THOPL_DICDI | 2.94e-28 | 1456 | 1701 | 394 | 610 | Thimet-like oligopeptidase OS=Dictyostelium discoideum OX=44689 GN=DDB_G0292362 PE=3 SV=1 |
| sp|Q8C1A5|THOP1_MOUSE | 7.36e-28 | 1443 | 1701 | 392 | 613 | Thimet oligopeptidase OS=Mus musculus OX=10090 GN=Thop1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001263 | 0.998697 | CS pos: 22-23. Pr: 0.9782 |