You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T27507-p1
You are here: Home > Sequence: FGRAMPH1_01T27507-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium graminearum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum | |||||||||||
| CAZyme ID | FGRAMPH1_01T27507-p1 | |||||||||||
| CAZy Family | GT69 | |||||||||||
| CAZyme Description | l-sorbosone dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 30 | 434 | 1.7e-176 | 0.9900249376558603 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 2.70e-18 | 1 | 426 | 1 | 385 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 271320 | NHL | 0.003 | 45 | 122 | 187 | 262 | NHL repeat unit of beta-propeller proteins. The NHL(NCL-1, HT2A and LIN-41)-repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. The repeats have a catalytic activity in Peptidyl-glycine alpha-amidating monooxygenase; proteolysis has shown that the Peptidyl-alpha-hydroxyglycine alpha-amidating lyase (PAL) activity is localized to the repeats. Tripartite motif-containing protein 32 interacts with the activation domain of Tat. This interaction is mediated by the NHL repeats. |
| 271331 | NHL_TRIM32_like | 0.003 | 47 | 123 | 195 | 267 | NHL repeat domain of the tripartite motif-containing protein 32 (TRIM32) and related proteins. The E3 ubiquitin-protein ligase TRIM32 (HT2A) is widely expressed and is responsible for ubiquinating a large variety of targets, including dysbindin (DTNBP1), NPHP7/Glis2, TAp73, and others. TRIM32 promotes disassociation of the plakoglobin-PI3K complex and reduces PI3K-Akt-FoxO signaling. Mutations in TRIM32 have been implemented in the two diverse diseases limb-girdle muscular dystrophy type 2H (LGMD2H) or sarcotubular myopathy (STM) and Bardet-Biedl syndrome type 11 (BBS11). The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 485 | 1 | 485 | |
| 0.0 | 1 | 485 | 1 | 485 | |
| 0.0 | 1 | 485 | 1 | 485 | |
| 0.0 | 1 | 485 | 1 | 485 | |
| 0.0 | 1 | 485 | 1 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.92e-151 | 26 | 437 | 1 | 405 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 2.21e-150 | 31 | 437 | 4 | 403 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 2.29e-150 | 31 | 437 | 5 | 404 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 7.80e-74 | 53 | 434 | 30 | 408 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.36e-17 | 101 | 426 | 158 | 433 | L-sorbosone dehydrogenase OS=Gluconacetobacter liquefaciens OX=89584 PE=4 SV=1 |
|
| 5.13e-08 | 51 | 438 | 47 | 378 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
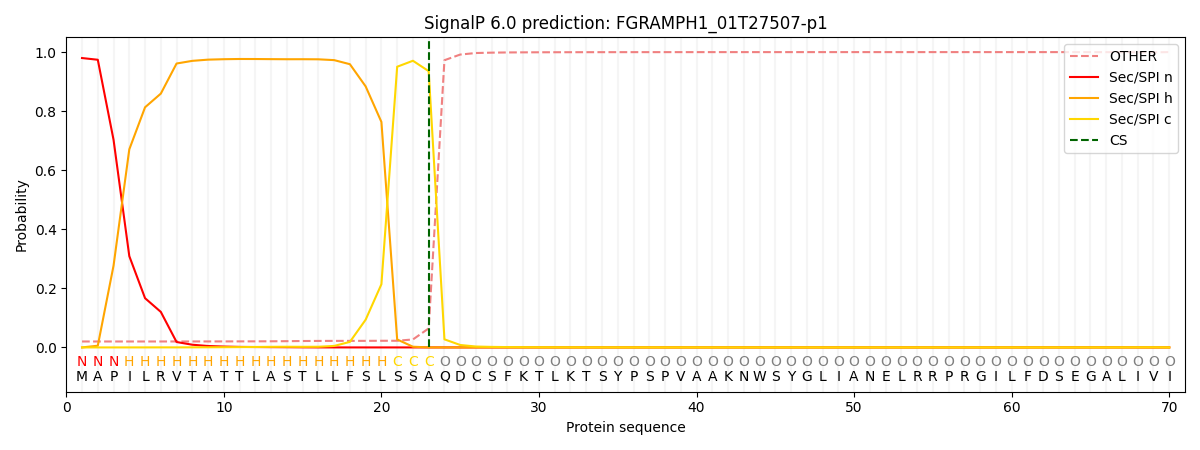
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.023675 | 0.976297 | CS pos: 23-24. Pr: 0.9355 |
