You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T25625-p1
You are here: Home > Sequence: FGRAMPH1_01T25625-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium graminearum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum | |||||||||||
| CAZyme ID | FGRAMPH1_01T25625-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | unnamed protein product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 75 | 246 | 5.7e-43 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 4.12e-59 | 75 | 235 | 1 | 160 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 494 | 1 | 494 | |
| 0.0 | 1 | 494 | 1 | 494 | |
| 2.06e-222 | 1 | 482 | 1 | 1029 | |
| 8.35e-158 | 1 | 255 | 1 | 255 | |
| 3.76e-154 | 1 | 246 | 1 | 250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.41e-36 | 71 | 243 | 74 | 243 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 6.63e-29 | 51 | 240 | 4 | 199 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 6.63e-29 | 51 | 240 | 4 | 199 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 6.63e-29 | 51 | 240 | 4 | 199 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 2.39e-28 | 51 | 240 | 4 | 199 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.46e-45 | 76 | 243 | 31 | 196 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 3.81e-43 | 57 | 246 | 22 | 213 | Cutinase CUT2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=CUT2 PE=1 SV=1 |
|
| 3.86e-41 | 76 | 243 | 31 | 195 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 7.51e-41 | 63 | 246 | 55 | 235 | Cutinase OS=Blumeria hordei OX=2867405 GN=CUT1 PE=3 SV=1 |
|
| 1.60e-38 | 76 | 247 | 32 | 201 | Cutinase pbc1 OS=Pyrenopeziza brassicae OX=76659 GN=pbc1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
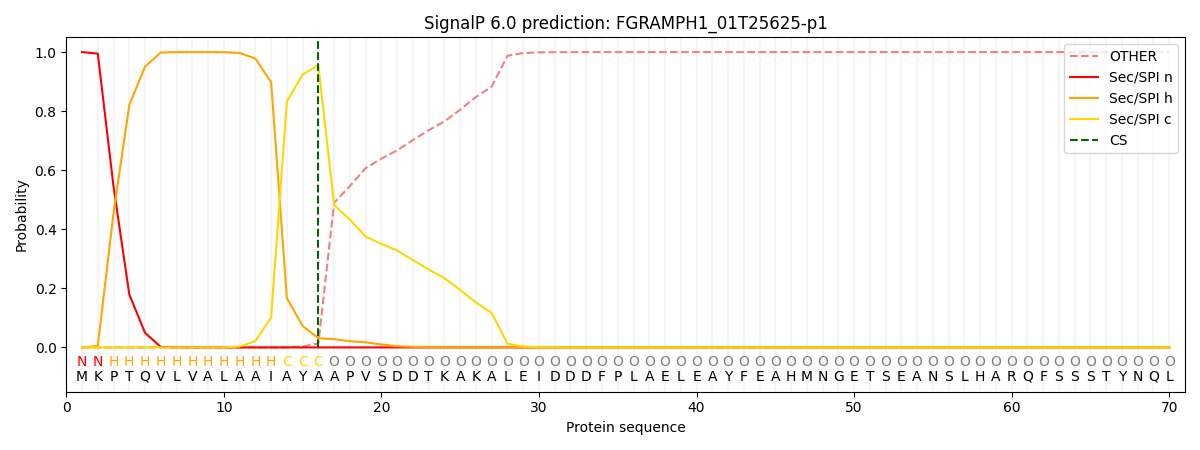
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000242 | 0.999746 | CS pos: 16-17. Pr: 0.9552 |
