You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T25415-p1
You are here: Home > Sequence: FGRAMPH1_01T25415-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium graminearum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum | |||||||||||
| CAZyme ID | FGRAMPH1_01T25415-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH142 | 30 | 503 | 1.1e-186 | 0.9603340292275574 |
| CBM13 | 677 | 815 | 5.9e-21 | 0.6968085106382979 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404972 | RicinB_lectin_2 | 1.51e-20 | 713 | 798 | 4 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| 404972 | RicinB_lectin_2 | 2.17e-14 | 670 | 751 | 3 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| 395611 | F5_F8_type_C | 2.61e-06 | 541 | 668 | 6 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| 404972 | RicinB_lectin_2 | 7.18e-06 | 760 | 814 | 4 | 58 | Ricin-type beta-trefoil lectin domain-like. |
| 395961 | Trehalase | 0.001 | 224 | 273 | 308 | 357 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 726 | 1 | 726 | |
| 0.0 | 1 | 726 | 1 | 726 | |
| 0.0 | 1 | 726 | 1 | 726 | |
| 0.0 | 12 | 726 | 1 | 708 | |
| 0.0 | 5 | 726 | 6 | 726 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.62e-110 | 13 | 503 | 616 | 1098 | Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
|
| 6.14e-106 | 13 | 503 | 616 | 1098 | Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
|
| 1.29e-23 | 35 | 506 | 305 | 809 | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.90e-32 | 35 | 676 | 336 | 1051 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
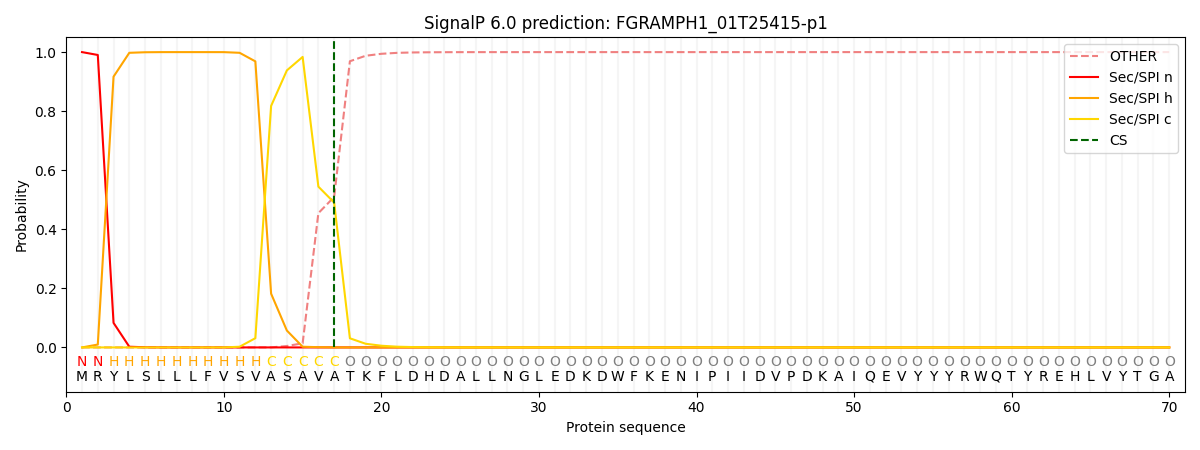
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000292 | 0.999680 | CS pos: 17-18. Pr: 0.4905 |
