You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T21485-p1
You are here: Home > Sequence: FGRAMPH1_01T21485-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium graminearum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum | |||||||||||
| CAZyme ID | FGRAMPH1_01T21485-p1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | pectin lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 110 | 295 | 1.3e-76 | 0.983957219251337 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 4.80e-44 | 109 | 298 | 3 | 190 | Amb_all domain. |
| 226384 | PelB | 6.16e-21 | 28 | 300 | 36 | 279 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 4.50e-20 | 119 | 294 | 29 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 411414 | MSCRAMM_ClfB | 1.04e-14 | 354 | 658 | 512 | 809 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411414 | MSCRAMM_ClfB | 2.15e-12 | 390 | 683 | 534 | 808 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 766 | 1 | 766 | |
| 0.0 | 1 | 766 | 1 | 766 | |
| 0.0 | 1 | 766 | 1 | 755 | |
| 0.0 | 1 | 766 | 1 | 745 | |
| 4.12e-308 | 1 | 766 | 1 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.84e-67 | 22 | 341 | 3 | 322 | Pectin Lyase B [Aspergillus niger] |
|
| 1.29e-60 | 22 | 339 | 3 | 319 | Pectin Lyase A [Aspergillus niger] |
|
| 3.42e-60 | 22 | 339 | 3 | 319 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 2.55e-08 | 711 | 746 | 1 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 4.77e-08 | 711 | 746 | 1 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.55e-90 | 15 | 418 | 16 | 401 | Probable pectin lyase F-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-2 PE=3 SV=1 |
|
| 8.01e-87 | 5 | 378 | 4 | 384 | Probable pectin lyase F-1 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-1 PE=3 SV=1 |
|
| 1.61e-83 | 9 | 394 | 12 | 389 | Probable pectin lyase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pelF PE=3 SV=1 |
|
| 2.34e-82 | 18 | 410 | 20 | 411 | Probable pectin lyase F OS=Aspergillus niger OX=5061 GN=pelF PE=3 SV=1 |
|
| 2.34e-82 | 18 | 410 | 20 | 411 | Probable pectin lyase F OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pelF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
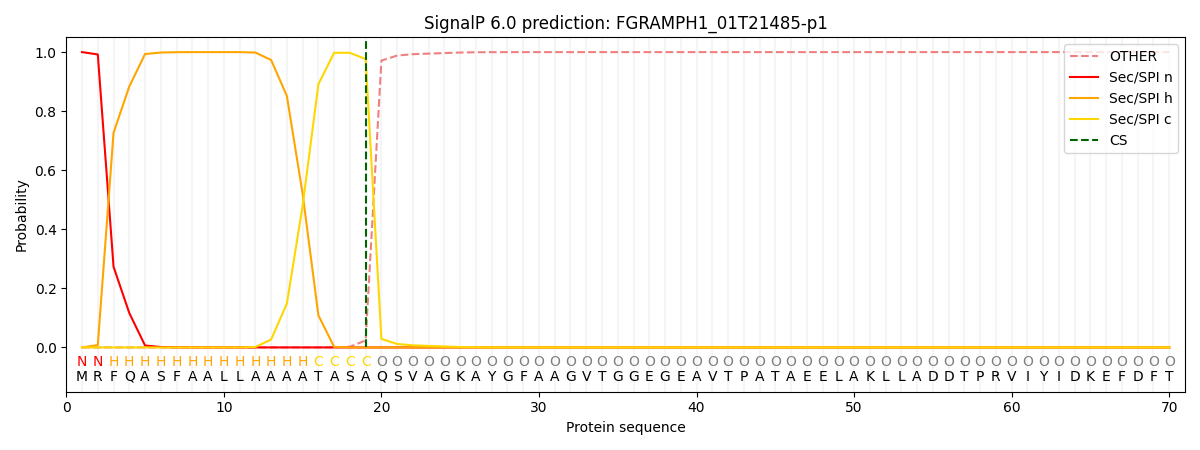
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000282 | 0.999677 | CS pos: 19-20. Pr: 0.9764 |
