You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T16129-p1
Basic Information
help
| Species |
Fusarium graminearum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum
|
| CAZyme ID |
FGRAMPH1_01T16129-p1
|
| CAZy Family |
GH3 |
| CAZyme Description |
alpha-L-rhamnosidase A
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 824 |
|
91401.96 |
5.3850 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FgraminearumPH-1 |
14145 |
229533 |
2 |
14143
|
|
| Gene Location |
No EC number prediction in FGRAMPH1_01T16129-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH78 |
382 |
794 |
3.8e-49 |
0.7321428571428571 |
FGRAMPH1_01T16129-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
824 |
1 |
824 |
| 0.0 |
1 |
824 |
1 |
824 |
| 0.0 |
11 |
824 |
8 |
819 |
| 0.0 |
1 |
820 |
1 |
803 |
| 0.0 |
1 |
820 |
1 |
803 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.05e-15 |
361 |
769 |
300 |
674 |
Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
FGRAMPH1_01T16129-p1 has no Swissprot hit.
This protein is predicted as OTHER
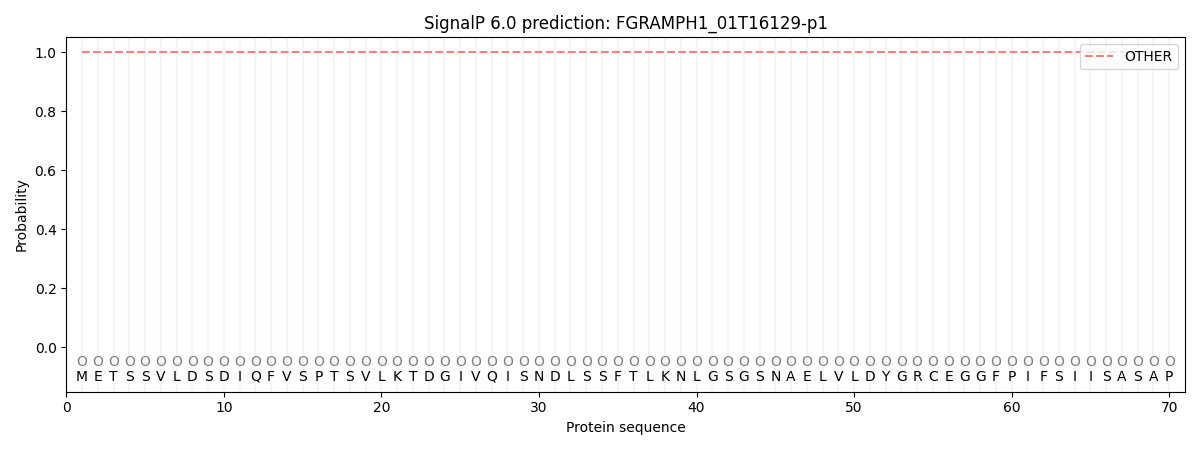
| Other |
SP_Sec_SPI |
CS Position |
| 1.000017 |
0.000049 |
|
There is no transmembrane helices in FGRAMPH1_01T16129-p1.

