You are browsing environment: FUNGIDB
FGRAMPH1_01T14827-p1
Basic Information
help
Species
Fusarium graminearum
Lineage
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum
CAZyme ID
FGRAMPH1_01T14827-p1
CAZy Family
GH20
CAZyme Description
f5 8 type c
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
316
HG970333|CGC31 34993.40
4.8754
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_FgraminearumPH-1
14145
229533
2
14143
Gene Location
No EC number prediction in FGRAMPH1_01T14827-p1.
Family
Start
End
Evalue
family coverage
GH128
43
280
2.8e-62
0.9776785714285714
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
371727
Glyco_hydro_cc
1.42e-74
35
280
5
235
Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
2.75e-243
1
316
1
316
2.75e-243
1
316
1
316
1.53e-240
1
316
1
316
2.84e-236
1
316
1
316
1.12e-222
1
310
1
310
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.25e-15
33
281
39
282
Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans]
more
FGRAMPH1_01T14827-p1 has no Swissprot hit.
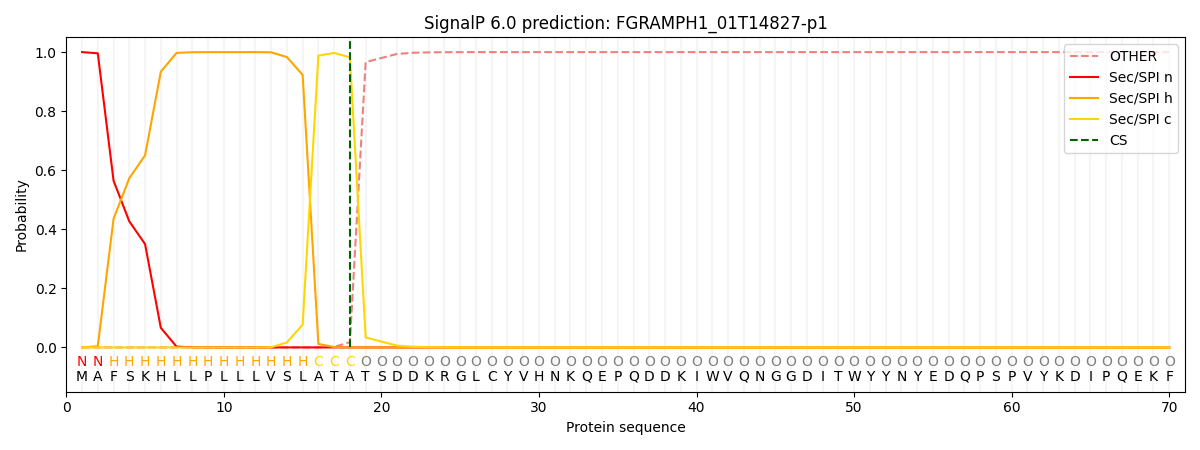
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000206
0.999764
CS pos: 18-19. Pr: 0.9822
There is no transmembrane helices in FGRAMPH1_01T14827-p1.