You are browsing environment: FUNGIDB
CAZyme Information: FGRAMPH1_01T13267-p1
You are here: Home > Sequence: FGRAMPH1_01T13267-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium graminearum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium graminearum | |||||||||||
| CAZyme ID | FGRAMPH1_01T13267-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | phosphoribosyl transferase domain | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 291 | 523 | 7.5e-40 | 0.462882096069869 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235028 | PRK02304 | 2.50e-22 | 955 | 1135 | 12 | 175 | adenine phosphoribosyltransferase; Provisional |
| 223577 | Apt | 1.66e-20 | 946 | 1122 | 5 | 169 | Adenine/guanine phosphoribosyltransferase or related PRPP-binding protein [Nucleotide transport and metabolism]. |
| 396238 | FAD_binding_4 | 2.97e-19 | 301 | 430 | 1 | 129 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 177930 | PLN02293 | 1.62e-17 | 946 | 1117 | 14 | 173 | adenine phosphoribosyltransferase |
| 223354 | GlcD | 8.80e-17 | 272 | 485 | 3 | 206 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.22e-13 | 301 | 736 | 63 | 483 | |
| 4.83e-12 | 301 | 554 | 164 | 402 | |
| 5.99e-11 | 298 | 736 | 60 | 482 | |
| 7.59e-10 | 256 | 733 | 16 | 491 | |
| 1.26e-09 | 301 | 740 | 61 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.77e-16 | 944 | 1134 | 2 | 178 | Crystal Structure of Human APRT wild type in complex with PRPP and Mg2+ [Homo sapiens],6FCH_B Crystal Structure of Human APRT wild type in complex with PRPP and Mg2+ [Homo sapiens],6FCL_A Crystal Structure of Human APRT wild type in complex with AMP [Homo sapiens],6FCL_B Crystal Structure of Human APRT wild type in complex with AMP [Homo sapiens],6HGP_A Crystal Structure of Human APRT wild type in complex with Phosphate ion. [Homo sapiens],6HGP_B Crystal Structure of Human APRT wild type in complex with Phosphate ion. [Homo sapiens],6HGR_A Crystal Structure of Human APRT wild type in complex with IMP [Homo sapiens],6HGR_B Crystal Structure of Human APRT wild type in complex with IMP [Homo sapiens],6HGS_A Crystal Structure of Human APRT wild type in complex with GMP [Homo sapiens],6HGS_B Crystal Structure of Human APRT wild type in complex with GMP [Homo sapiens] |
|
| 2.84e-16 | 944 | 1134 | 3 | 179 | Crystal Structure of Human APRT wild type in complex with Adenine, PRPP and Mg2+ [Homo sapiens],6FCI_B Crystal Structure of Human APRT wild type in complex with Adenine, PRPP and Mg2+ [Homo sapiens],6FCI_C Crystal Structure of Human APRT wild type in complex with Adenine, PRPP and Mg2+ [Homo sapiens],6FCI_D Crystal Structure of Human APRT wild type in complex with Adenine, PRPP and Mg2+ [Homo sapiens],6HGQ_A Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ [Homo sapiens],6HGQ_B Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ [Homo sapiens],6HGQ_C Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ [Homo sapiens],6HGQ_D Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ [Homo sapiens] |
|
| 2.90e-16 | 944 | 1134 | 4 | 180 | Human Adenine Phosphoribosyltransferase [Homo sapiens],1ZN7_A Human Adenine Phosphoribosyltransferase Complexed with PRPP, ADE and R5P [Homo sapiens],1ZN7_B Human Adenine Phosphoribosyltransferase Complexed with PRPP, ADE and R5P [Homo sapiens],1ZN8_A Human Adenine Phosphoribosyltransferase Complexed with AMP, in Space Group P1 at 1.76 A Resolution [Homo sapiens],1ZN8_B Human Adenine Phosphoribosyltransferase Complexed with AMP, in Space Group P1 at 1.76 A Resolution [Homo sapiens],1ZN9_A Human Adenine Phosphoribosyltransferase in Apo and AMP Complexed Forms [Homo sapiens],1ZN9_B Human Adenine Phosphoribosyltransferase in Apo and AMP Complexed Forms [Homo sapiens] |
|
| 2.90e-16 | 944 | 1134 | 4 | 180 | Crystal Structure of Mutant R89Q of human Adenine phosphoribosyltransferase [Homo sapiens] |
|
| 3.95e-16 | 944 | 1117 | 4 | 168 | Crystal Structure of F173G Mutant of Human APRT [Homo sapiens],4X45_B Crystal Structure of F173G Mutant of Human APRT [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.20e-27 | 299 | 733 | 46 | 437 | FAD-linked oxidoreductase DDB_G0289697 OS=Dictyostelium discoideum OX=44689 GN=DDB_G0289697 PE=2 SV=1 |
|
| 4.81e-18 | 949 | 1122 | 5 | 166 | Adenine phosphoribosyltransferase OS=Roseiflexus castenholzii (strain DSM 13941 / HLO8) OX=383372 GN=apt PE=3 SV=1 |
|
| 5.89e-18 | 945 | 1134 | 5 | 180 | Adenine phosphoribosyltransferase OS=Mastomys natalensis OX=10112 GN=APRT PE=3 SV=1 |
|
| 8.01e-18 | 945 | 1134 | 5 | 180 | Adenine phosphoribosyltransferase OS=Mus spicilegus OX=10103 GN=Aprt PE=3 SV=1 |
|
| 8.01e-18 | 944 | 1134 | 4 | 180 | Adenine phosphoribosyltransferase OS=Mus pahari OX=10093 GN=Aprt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
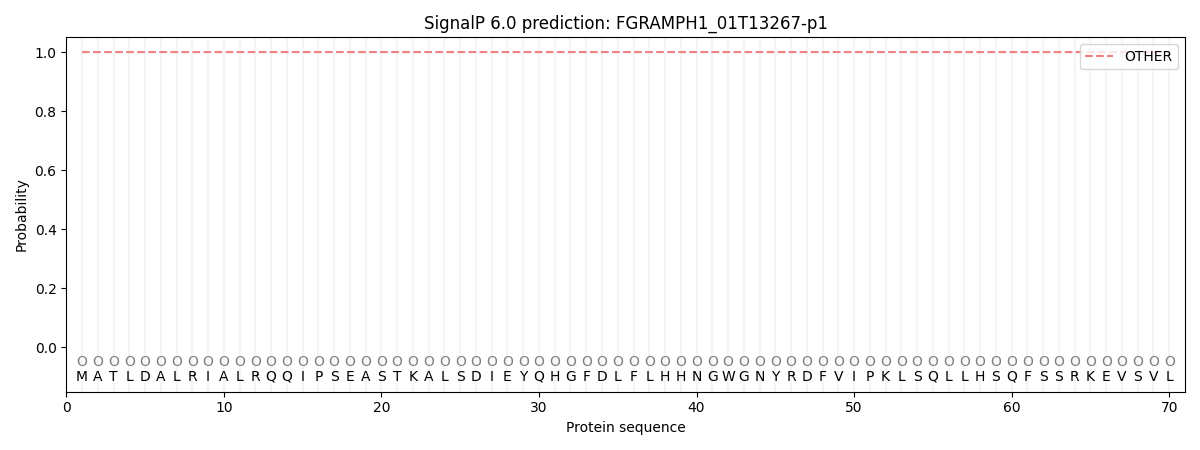
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000057 | 0.000000 |
