You are browsing environment: FUNGIDB
CAZyme Information: FFUJ_12620-t31_1-p1
You are here: Home > Sequence: FFUJ_12620-t31_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium fujikuroi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium fujikuroi | |||||||||||
| CAZyme ID | FFUJ_12620-t31_1-p1 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | related to endo-1,4-beta-xylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:45 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 29 | 325 | 9.2e-102 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 4.67e-149 | 29 | 324 | 1 | 310 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 7.06e-121 | 73 | 322 | 2 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 1.52e-77 | 63 | 323 | 58 | 338 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 0.006 | 74 | 103 | 34 | 63 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.24e-243 | 1 | 327 | 1 | 327 | |
| 5.35e-242 | 1 | 327 | 1 | 327 | |
| 3.09e-241 | 1 | 327 | 1 | 327 | |
| 3.09e-241 | 1 | 327 | 1 | 327 | |
| 1.78e-238 | 1 | 327 | 1 | 328 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.77e-128 | 25 | 326 | 2 | 302 | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
|
| 7.86e-128 | 25 | 327 | 2 | 302 | The xylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
|
| 1.53e-127 | 25 | 326 | 2 | 302 | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH 1,2-(4-DEOXY-BETA-L-THREO-HEX-4-ENOPYRANOSYLURONIC ACID)-BETA-1,4-XYLOTRIOSE) [Penicillium simplicissimum],1B31_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, NATIVE WITH PEG200 AS CRYOPROTECTANT [Penicillium simplicissimum],1B3V_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOSE [Penicillium simplicissimum],1B3W_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOBIOSE [Penicillium simplicissimum],1B3X_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTRIOSE [Penicillium simplicissimum],1B3Y_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTETRAOSE [Penicillium simplicissimum],1B3Z_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOPENTAOSE [Penicillium simplicissimum],1BG4_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM [Penicillium simplicissimum] |
|
| 3.18e-127 | 24 | 327 | 1 | 302 | Structural studies on the mobility in the active site of the Thermoascus aurantiacus xylanase I [Thermoascus aurantiacus] |
|
| 4.52e-127 | 25 | 327 | 2 | 302 | 0.89A Ultra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.31e-179 | 1 | 327 | 1 | 327 | Endo-1,4-beta-xylanase C OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLC PE=1 SV=1 |
|
| 2.34e-136 | 2 | 326 | 6 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
|
| 1.17e-135 | 1 | 327 | 1 | 331 | Endo-1,4-beta-xylanase 2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL2 PE=3 SV=1 |
|
| 1.44e-135 | 10 | 326 | 14 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
|
| 8.76e-132 | 2 | 326 | 6 | 316 | Probable endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
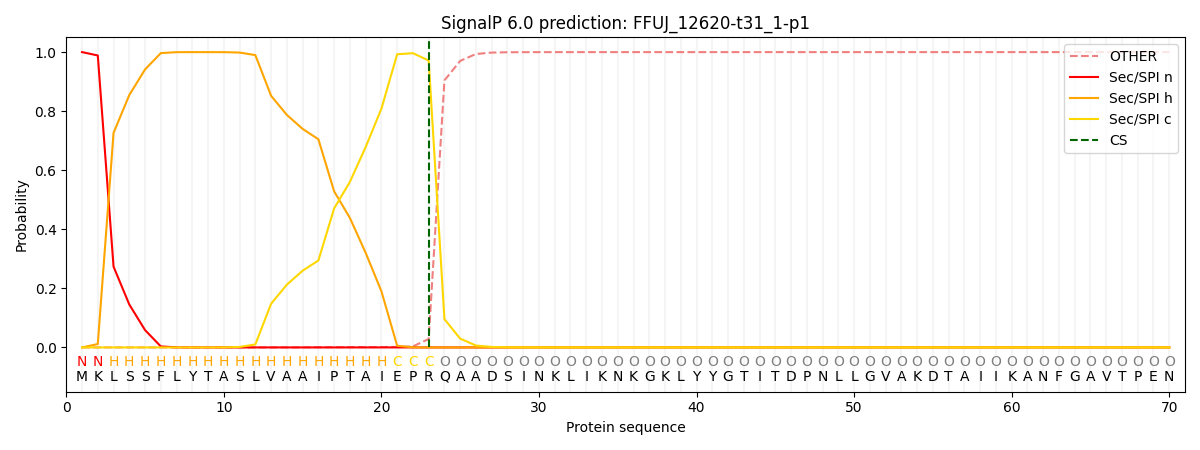
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000474 | 0.999485 | CS pos: 23-24. Pr: 0.9714 |
