You are browsing environment: FUNGIDB
CAZyme Information: FFUJ_06613-t31_1-p1
You are here: Home > Sequence: FFUJ_06613-t31_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium fujikuroi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium fujikuroi | |||||||||||
| CAZyme ID | FFUJ_06613-t31_1-p1 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | uncharacterized protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH142 | 32 | 505 | 1.3e-171 | 0.9624217118997912 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397353 | Glyco_hydro_63 | 3.94e-05 | 294 | 432 | 361 | 489 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| 225942 | GDB1 | 0.004 | 237 | 354 | 444 | 541 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 670 | 1 | 670 | |
| 0.0 | 1 | 670 | 1 | 670 | |
| 0.0 | 1 | 670 | 1 | 670 | |
| 0.0 | 1 | 670 | 1 | 670 | |
| 1.66e-274 | 25 | 669 | 24 | 677 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.63e-79 | 34 | 506 | 651 | 1101 | Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
|
| 1.08e-75 | 34 | 506 | 651 | 1101 | Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
|
| 3.92e-19 | 36 | 552 | 305 | 856 | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.87e-18 | 36 | 552 | 336 | 887 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
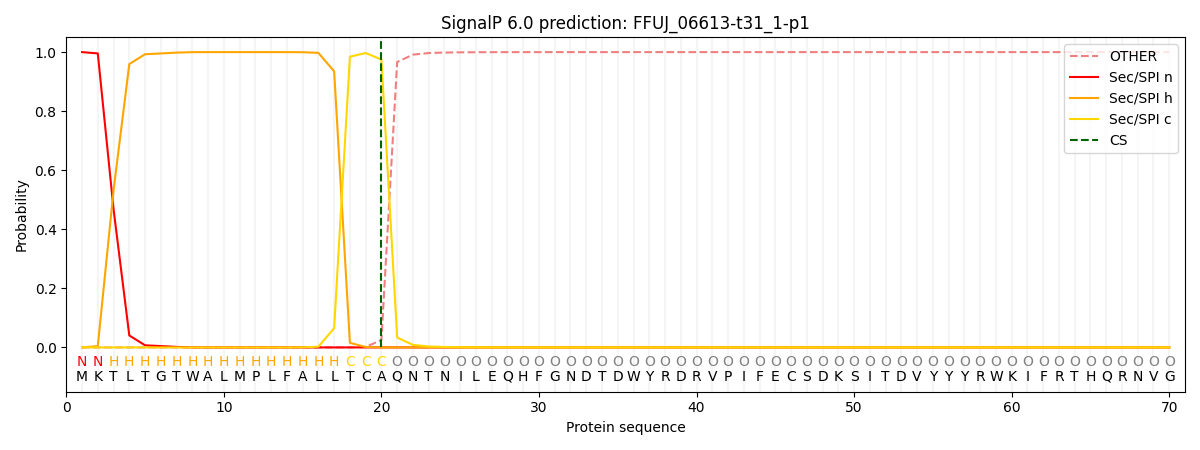
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000246 | 0.999735 | CS pos: 20-21. Pr: 0.9749 |
