You are browsing environment: FUNGIDB
CAZyme Information: FFUJ_01901-t31_1-p1
You are here: Home > Sequence: FFUJ_01901-t31_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium fujikuroi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium fujikuroi | |||||||||||
| CAZyme ID | FFUJ_01901-t31_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | related to beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 163 | 621 | 1.2e-101 | 0.9883449883449883 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225343 | BglB | 2.95e-82 | 162 | 610 | 2 | 443 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 395176 | Glyco_hydro_1 | 2.26e-81 | 164 | 621 | 5 | 452 | Glycosyl hydrolase family 1. |
| 215539 | PLN02998 | 2.67e-38 | 163 | 598 | 30 | 464 | beta-glucosidase |
| 184102 | PRK13511 | 3.80e-38 | 162 | 609 | 3 | 455 | 6-phospho-beta-galactosidase; Provisional |
| 215435 | PLN02814 | 4.63e-38 | 163 | 605 | 27 | 466 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 621 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.81e-61 | 163 | 609 | 21 | 471 | Chain A, Beta-glucosidase 42 [Arabidopsis thaliana] |
|
| 2.66e-61 | 165 | 609 | 10 | 451 | Chain A, Beta-glucosidase [Phanerodontia chrysosporium],2E3Z_B Chain B, Beta-glucosidase [Phanerodontia chrysosporium],2E40_A Chain A, Beta-glucosidase [Phanerodontia chrysosporium],2E40_B Chain B, Beta-glucosidase [Phanerodontia chrysosporium] |
|
| 1.20e-60 | 162 | 622 | 10 | 473 | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Tris [Neotermes koshunensis],3VIF_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with gluconolactone [Neotermes koshunensis],3VIG_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with 1-deoxynojirimycin [Neotermes koshunensis],3VIH_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with glycerol [Neotermes koshunensis],3VII_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with Bis-Tris [Neotermes koshunensis] |
|
| 3.22e-60 | 162 | 622 | 10 | 473 | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with para-nitrophenyl-beta-D-glucopyranoside [Neotermes koshunensis],3VIM_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product [Neotermes koshunensis],3VIN_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product [Neotermes koshunensis],3VIO_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product [Neotermes koshunensis],3VIP_A Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product [Neotermes koshunensis] |
|
| 5.87e-60 | 164 | 609 | 89 | 560 | Chain A, Beta-hexosyltransferase [Hamamotoa singularis],7L74_B Chain B, Beta-hexosyltransferase [Hamamotoa singularis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.12e-61 | 163 | 609 | 20 | 470 | Beta-glucosidase 42 OS=Arabidopsis thaliana OX=3702 GN=BGLU42 PE=2 SV=1 |
|
| 1.08e-60 | 163 | 609 | 14 | 462 | Beta-glucosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU4 PE=2 SV=1 |
|
| 1.28e-60 | 165 | 609 | 7 | 448 | Beta-glucosidase 1A OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1A PE=1 SV=1 |
|
| 2.58e-56 | 165 | 609 | 12 | 463 | Beta-glucosidase 1B OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1B PE=1 SV=1 |
|
| 3.66e-55 | 163 | 608 | 36 | 496 | Beta-glucosidase 40 OS=Arabidopsis thaliana OX=3702 GN=BGLU40 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
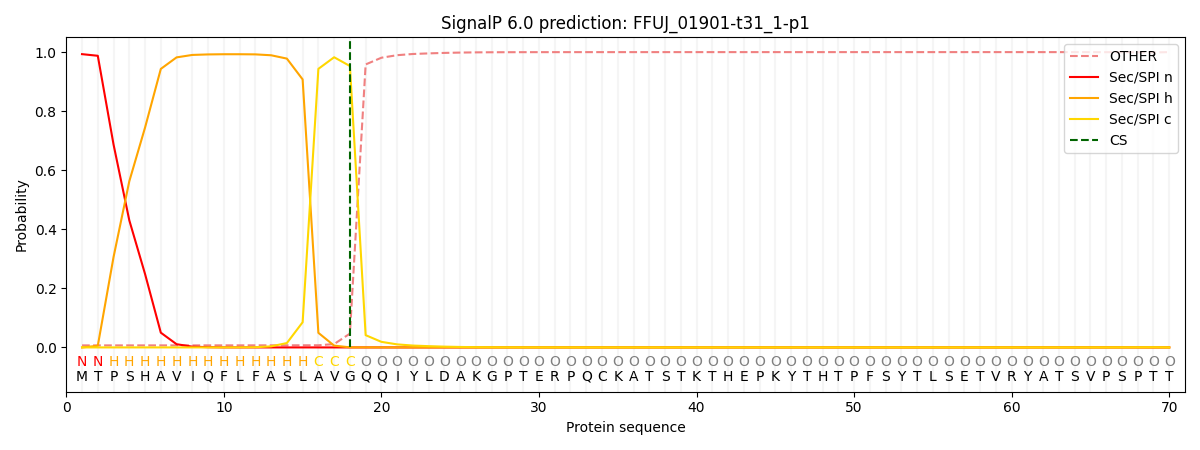
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.011586 | 0.988375 | CS pos: 18-19. Pr: 0.9529 |
