You are browsing environment: FUNGIDB
CAZyme Information: FFUJ_00351-t31_1-p1
You are here: Home > Sequence: FFUJ_00351-t31_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium fujikuroi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium fujikuroi | |||||||||||
| CAZyme ID | FFUJ_00351-t31_1-p1 | |||||||||||
| CAZy Family | AA16 | |||||||||||
| CAZyme Description | probable galactose oxidase precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.3.9:34 | 1.1.3.13:3 | 1.1.3.-:1 | 1.1.3.7:1 | 1.1.3.47:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA5 | 444 | 1056 | 1.5e-241 | 0.9948979591836735 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 199882 | E_set_GO_C | 2.96e-31 | 957 | 1056 | 1 | 103 | C-terminal Early set domain associated with the catalytic domain of galactose oxidase. E or "early" set domains are associated with the catalytic domain of galactose oxidase at the C-terminal end. Galactose oxidase is an extracellular monomeric enzyme which catalyzes the stereospecific oxidation of a broad range of primary alcohol substrates and possesses a unique mononuclear copper site essential for catalyzing a two-electron transfer reaction during the oxidation of primary alcohols to corresponding aldehydes. The second redox active center necessary for the reaction was found to be situated at a tyrosine residue. The C-terminal domain of galactose oxidase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| 401164 | DUF1929 | 1.10e-27 | 968 | 1056 | 1 | 91 | Domain of unknown function (DUF1929). Members of this family adopt a secondary structure consisting of a bundle of seven, mostly antiparallel, beta-strands surrounding a hydrophobic core. The 7 strands are arranged in 2 sheets, in a Greek-key topology. Their precise function, has not, as yet, been defined, though they are mostly found in sugar-utilising enzymes, such as galactose oxidase. |
| 395611 | F5_F8_type_C | 7.21e-22 | 434 | 561 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| 238014 | FA58C | 2.79e-19 | 433 | 552 | 12 | 134 | Substituted updates: Jan 31, 2002 |
| 214572 | FA58C | 1.24e-07 | 422 | 554 | 1 | 130 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1059 | 1 | 1059 | |
| 0.0 | 1 | 1059 | 1 | 1059 | |
| 0.0 | 1 | 1059 | 1 | 1059 | |
| 0.0 | 1 | 1059 | 1 | 1072 | |
| 0.0 | 1 | 1058 | 1 | 1067 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 419 | 1057 | 23 | 660 | Chain A, GALACTOSE OXIDASE [Fusarium graminearum] |
|
| 0.0 | 419 | 1057 | 1 | 638 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 0.0 | 419 | 1057 | 1 | 638 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 0.0 | 419 | 1057 | 1 | 638 | Chain A, Galactose oxidase [Fusarium graminearum] |
|
| 0.0 | 419 | 1057 | 1 | 638 | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 415 | 1057 | 38 | 679 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
|
| 0.0 | 415 | 1057 | 38 | 679 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
|
| 7.88e-18 | 428 | 561 | 504 | 641 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
|
| 1.86e-15 | 849 | 1056 | 387 | 614 | Aldehyde oxidase GLOX1 OS=Arabidopsis thaliana OX=3702 GN=GLOX1 PE=2 SV=1 |
|
| 3.86e-13 | 402 | 568 | 20 | 188 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
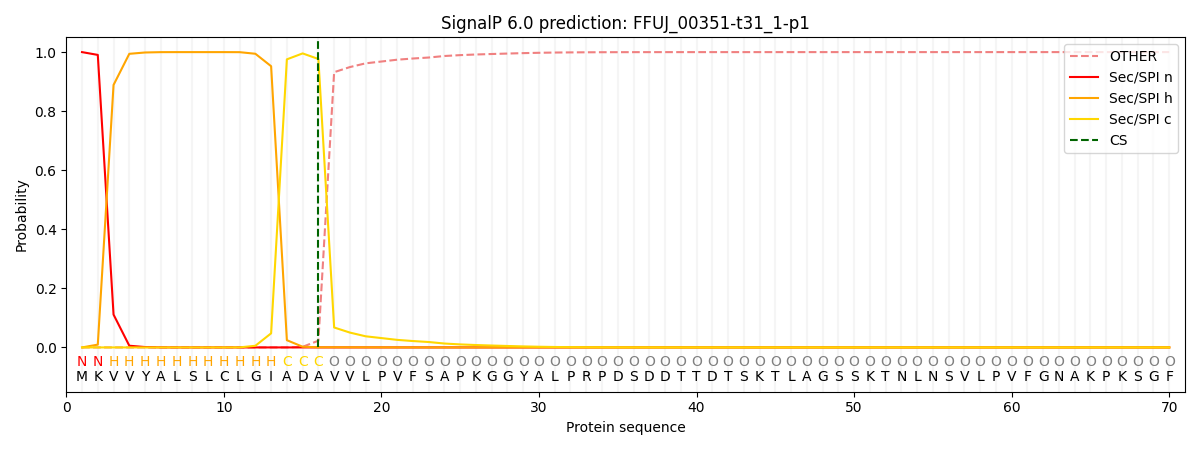
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000283 | 0.999687 | CS pos: 16-17. Pr: 0.9767 |
