You are browsing environment: FUNGIDB
CAZyme Information: EXJ73669.1
You are here: Home > Sequence: EXJ73669.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cladophialophora psammophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora psammophila | |||||||||||
| CAZyme ID | EXJ73669.1 | |||||||||||
| CAZy Family | GT15 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 212 | 460 | 1.1e-19 | 0.954983922829582 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 3.31e-41 | 141 | 460 | 1 | 301 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 223039 | PHA03307 | 1.51e-04 | 89 | 186 | 106 | 215 | transcriptional regulator ICP4; Provisional |
| 237057 | PRK12323 | 4.83e-04 | 89 | 186 | 397 | 488 | DNA polymerase III subunit gamma/tau. |
| 236776 | PRK10856 | 6.00e-04 | 94 | 189 | 154 | 252 | cytoskeleton protein RodZ. |
| 177464 | PHA02682 | 6.51e-04 | 89 | 179 | 84 | 161 | ORF080 virion core protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.27e-56 | 207 | 462 | 138 | 391 | |
| 5.20e-55 | 205 | 460 | 149 | 407 | |
| 2.55e-54 | 211 | 462 | 110 | 363 | |
| 2.55e-54 | 211 | 459 | 110 | 360 | |
| 2.55e-54 | 211 | 459 | 110 | 360 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.65e-43 | 207 | 459 | 133 | 385 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 5.09e-43 | 207 | 460 | 123 | 376 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 4.35e-40 | 207 | 459 | 130 | 382 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 1.12e-30 | 213 | 459 | 284 | 537 | Probable family 17 glucosidase SCW11 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW11 PE=1 SV=1 |
|
| 3.17e-28 | 206 | 458 | 304 | 559 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
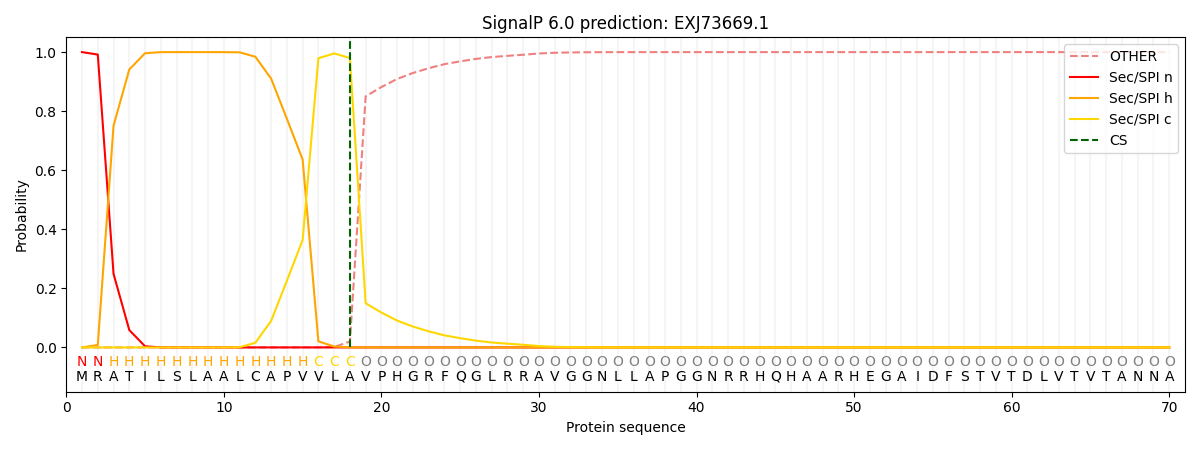
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000244 | 0.999727 | CS pos: 18-19. Pr: 0.9799 |
