You are browsing environment: FUNGIDB
CAZyme Information: EXJ73064.1
You are here: Home > Sequence: EXJ73064.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cladophialophora psammophila | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora psammophila | |||||||||||
| CAZyme ID | EXJ73064.1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | Cellulase domain-containing protein [Source:UniProtKB/TrEMBL;Acc:W9XRZ4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 68 | 368 | 3.3e-95 | 0.9931740614334471 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 9.76e-19 | 17 | 292 | 15 | 264 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 0.001 | 73 | 269 | 19 | 195 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.80e-177 | 9 | 547 | 6 | 505 | |
| 2.60e-170 | 9 | 547 | 11 | 515 | |
| 8.27e-170 | 7 | 547 | 6 | 508 | |
| 2.75e-160 | 9 | 542 | 42 | 524 | |
| 2.91e-150 | 9 | 543 | 35 | 519 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.91e-55 | 12 | 449 | 6 | 412 | Glucan 1,3-beta-glucosidase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=exg3 PE=3 SV=1 |
|
| 1.86e-49 | 14 | 438 | 40 | 441 | Uncharacterized glycosyl hydrolase YBR056W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YBR056W PE=1 SV=1 |
|
| 2.34e-20 | 22 | 370 | 44 | 392 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
|
| 2.34e-20 | 22 | 370 | 44 | 392 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
|
| 3.80e-20 | 22 | 370 | 33 | 382 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
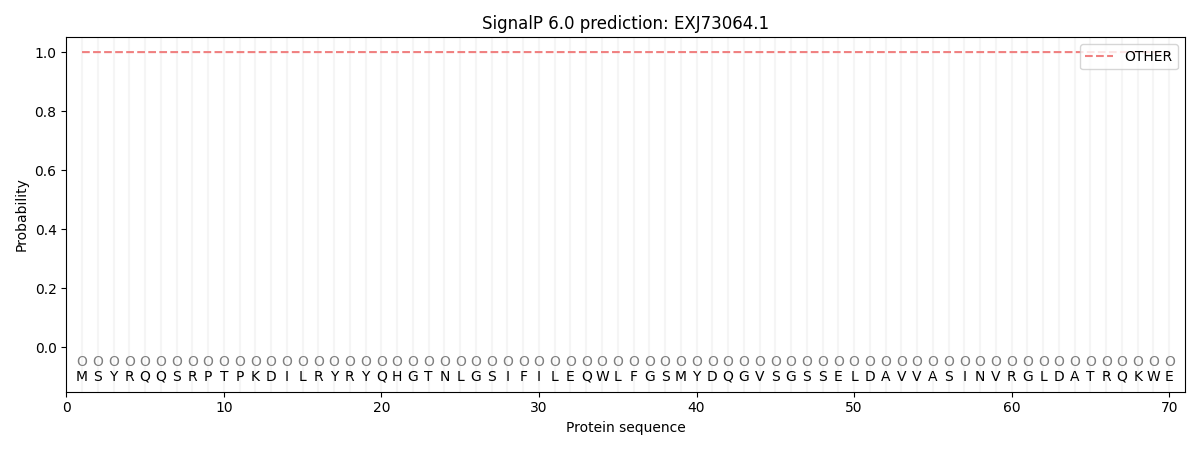
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000043 | 0.000001 |
