You are browsing environment: FUNGIDB
EXJ69370.1
Basic Information
help
Species
Cladophialophora psammophila
Lineage
Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora psammophila
CAZyme ID
EXJ69370.1
CAZy Family
GH17
CAZyme Description
unspecified product
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
426
44037.96
4.3690
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CpsammophilaCBS110553
13421
1182543
0
13421
Gene Location
No EC number prediction in EXJ69370.1.
Family
Start
End
Evalue
family coverage
AA14
19
287
1.3e-82
0.9882352941176471
EXJ69370.1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
6.34e-130
10
287
9
286
3.39e-127
10
287
9
286
1.50e-124
10
286
9
285
3.37e-122
10
286
9
285
5.32e-119
10
286
9
286
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5.61e-46
19
286
1
261
Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina],5NO7_B Crystal Structure of a Xylan-active Lytic Polysaccharide Monooxygenase from Pycnoporus coccineus. [Trametes cinnabarina]
more
EXJ69370.1 has no Swissprot hit.
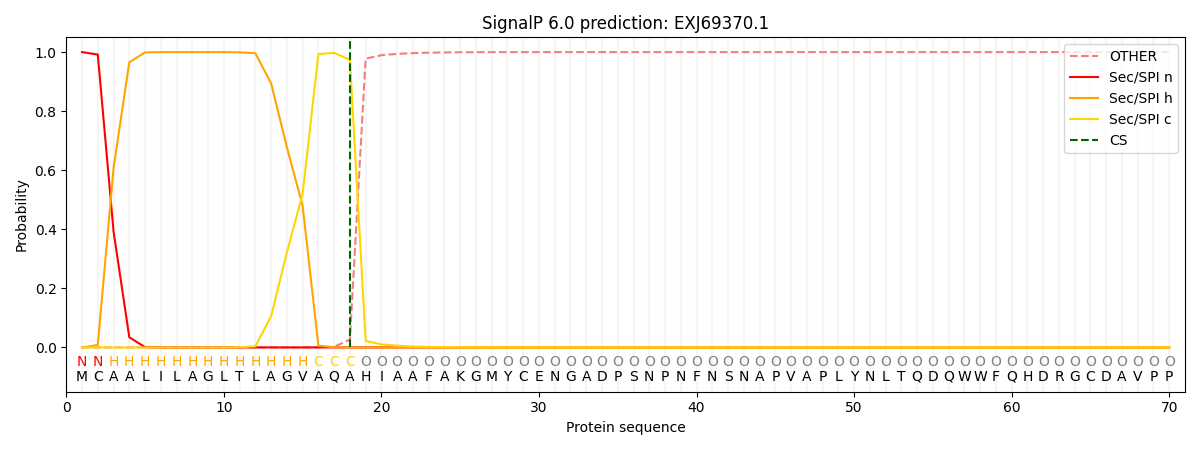
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000212
0.999755
CS pos: 18-19. Pr: 0.9734
There is no transmembrane helices in EXJ69370.1.