You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000023537-p1
You are here: Home > Sequence: EPrPVT00000023537-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000023537-p1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | Pectin lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 130 | 296 | 4.4e-50 | 0.93048128342246 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 2.44e-21 | 130 | 296 | 4 | 177 | Amb_all domain. |
| 226384 | PelB | 6.82e-10 | 103 | 282 | 51 | 248 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 6.92e-04 | 144 | 296 | 32 | 202 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 380308 | EmaA_autotrans | 0.002 | 106 | 227 | 1415 | 1550 | collagen-binding adhesin autotransporter EmaA. EmaA (extracellular matrix protein adhesin ) is an outer membrane protein first described in Aggregatibacter (Actinobacillus) actinomycetemcomitans, a member of the Gammaproteobacteria. It is a glycoprotein, and has a C-terminal beta-barrel domain that marks it as an autotransporter. It serves as an adhesin that binds collagen, the most abundant material in the host extracellular matrix. It shares homology with the collagen-binding protein YadA of Yersinia enterocolitica. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.73e-119 | 28 | 374 | 22 | 400 | |
| 1.06e-118 | 28 | 374 | 22 | 400 | |
| 1.46e-113 | 14 | 341 | 8 | 368 | |
| 7.29e-109 | 49 | 341 | 134 | 459 | |
| 1.16e-106 | 13 | 365 | 7 | 396 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.06e-53 | 31 | 335 | 5 | 325 | Pectin Lyase A [Aspergillus niger] |
|
| 3.52e-51 | 31 | 335 | 5 | 325 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 8.34e-48 | 30 | 331 | 4 | 322 | Pectin Lyase B [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.19e-61 | 31 | 344 | 24 | 361 | Probable pectin lyase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelB PE=3 SV=2 |
|
| 7.51e-59 | 11 | 331 | 4 | 340 | Pectin lyase 2 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pel2 PE=1 SV=1 |
|
| 7.51e-59 | 11 | 331 | 4 | 340 | Probable pectin lyase D OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pelD PE=3 SV=1 |
|
| 3.27e-57 | 14 | 351 | 8 | 373 | Probable pectin lyase F OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pelF PE=3 SV=1 |
|
| 3.27e-57 | 14 | 351 | 8 | 373 | Probable pectin lyase F OS=Aspergillus niger OX=5061 GN=pelF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
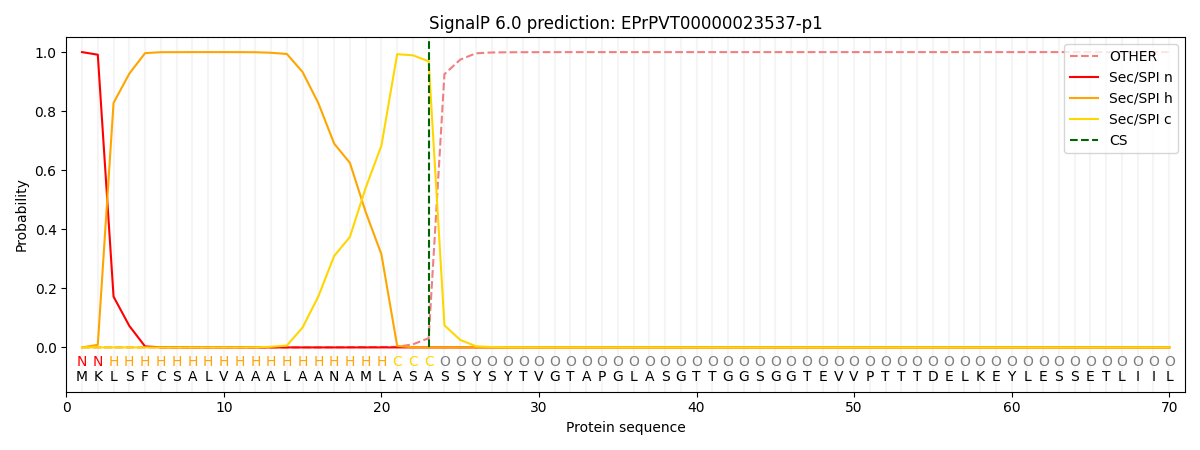
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000456 | 0.999507 | CS pos: 23-24. Pr: 0.9687 |
