You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000022908-p1
You are here: Home > Sequence: EPrPVT00000022908-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000022908-p1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | Glycoside hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:71 | 3.2.1.-:11 | 3.2.1.38:3 | 3.2.1.23:2 | 3.2.1.74:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 29 | 214 | 1.1e-75 | 0.4219114219114219 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274539 | BGL | 9.53e-81 | 31 | 212 | 1 | 178 | beta-galactosidase. |
| 225343 | BglB | 8.04e-77 | 30 | 212 | 4 | 186 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 395176 | Glyco_hydro_1 | 3.09e-75 | 30 | 211 | 5 | 182 | Glycosyl hydrolase family 1. |
| 184102 | PRK13511 | 4.76e-59 | 30 | 214 | 5 | 180 | 6-phospho-beta-galactosidase; Provisional |
| 215435 | PLN02814 | 6.70e-55 | 30 | 212 | 28 | 202 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.93e-99 | 24 | 218 | 26 | 220 | |
| 3.61e-94 | 29 | 218 | 12 | 201 | |
| 6.98e-83 | 24 | 211 | 17 | 204 | |
| 7.33e-56 | 30 | 211 | 39 | 220 | |
| 7.33e-56 | 30 | 211 | 39 | 220 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-55 | 30 | 211 | 11 | 188 | Chain A, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1H_B Chain B, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1M_A Chain A, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_B Chain B, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_C Chain C, Ancestral reconstructed glycosidase [synthetic construct] |
|
| 1.11e-55 | 28 | 218 | 41 | 231 | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF6_B Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF7_A Structure of Strictosidine Glucosidase [Rauvolfia serpentina],2JF7_B Structure of Strictosidine Glucosidase [Rauvolfia serpentina],3ZJ7_A Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ7_B Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ8_A Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina],3ZJ8_B Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina] |
|
| 1.02e-54 | 30 | 211 | 45 | 221 | Crystal structure of psychrophilic beta-glucosidase BglU from Micrococcus antarcticus [Micrococcus antarcticus] |
|
| 2.03e-53 | 30 | 212 | 12 | 189 | Chain A, Beta-glucosidase B [Paenibacillus polymyxa] |
|
| 2.11e-53 | 30 | 212 | 14 | 191 | Beta-Glucosidase B From Bacillus Polymyxa Complexed With 2- F-Glucose [Paenibacillus polymyxa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.18e-56 | 28 | 211 | 72 | 255 | Furcatin hydrolase OS=Viburnum furcatum OX=237940 PE=1 SV=1 |
|
| 5.69e-55 | 28 | 218 | 41 | 231 | Strictosidine-O-beta-D-glucosidase OS=Rauvolfia serpentina OX=4060 GN=SGR1 PE=1 SV=1 |
|
| 3.41e-53 | 30 | 211 | 35 | 214 | Putative beta-glucosidase 41 OS=Arabidopsis thaliana OX=3702 GN=BGLU41 PE=3 SV=2 |
|
| 9.59e-53 | 30 | 212 | 8 | 185 | Beta-glucosidase B OS=Paenibacillus polymyxa OX=1406 GN=bglB PE=1 SV=1 |
|
| 1.62e-52 | 30 | 211 | 44 | 224 | Beta-D-glucopyranosyl abscisate beta-glucosidase OS=Arabidopsis thaliana OX=3702 GN=BGLU18 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
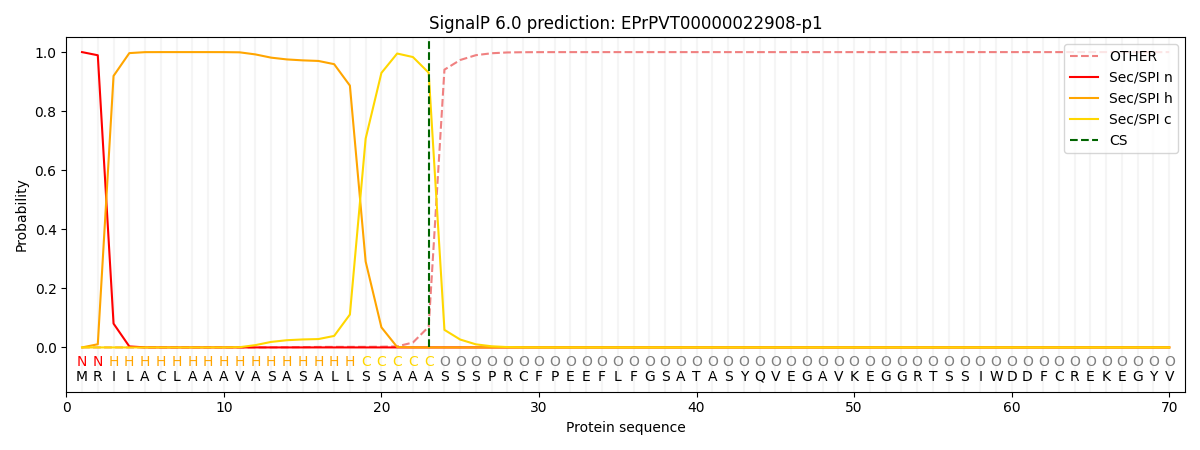
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000278 | 0.999697 | CS pos: 23-24. Pr: 0.9302 |
