You are browsing environment: FUNGIDB
CAZyme Information: EPrPVT00000022331-p1
You are here: Home > Sequence: EPrPVT00000022331-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
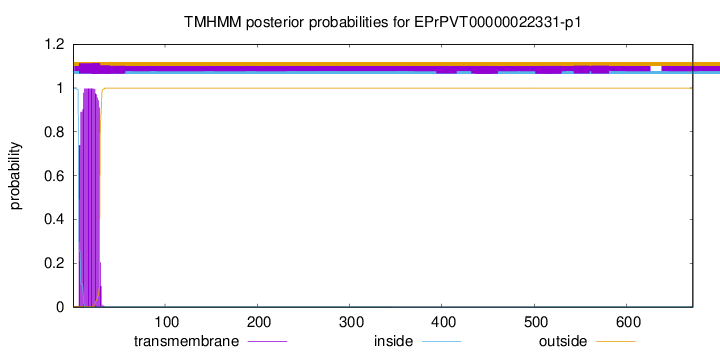
TMHMM annotations
Basic Information help
| Species | Phytopythium vexans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Phytopythium; Phytopythium vexans | |||||||||||
| CAZyme ID | EPrPVT00000022331-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | DEHA2C12078p | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 250 | 548 | 5.6e-42 | 0.9848484848484849 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 9.23e-25 | 250 | 549 | 2 | 272 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
| 173412 | PTZ00121 | 2.82e-05 | 93 | 248 | 1620 | 1801 | MAEBL; Provisional |
| 411258 | pilus_FilE | 3.45e-05 | 93 | 234 | 39 | 189 | putative pilus assembly protein FilE. FilE is found almost exclusively in the genus Acinetobacter, and is assigned as a putative pilus system protein from local genomic contexts that include several additional putative pilus system proteins. Note that some members of this protein family have proline-rich repeat regions for which spurious translation in another reading frame can give a false-positive match to Pfam's collagen repeat region HMM, PF01391. |
| 411408 | PspC_subgroup_2 | 3.90e-05 | 94 | 258 | 317 | 480 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| 411407 | PspC_subgroup_1 | 1.25e-04 | 91 | 249 | 164 | 332 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.46e-121 | 243 | 636 | 105 | 502 | |
| 5.47e-104 | 242 | 633 | 136 | 526 | |
| 1.68e-91 | 250 | 633 | 39 | 431 | |
| 1.34e-84 | 242 | 628 | 140 | 493 | |
| 7.05e-70 | 250 | 623 | 79 | 457 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.21e-13 | 244 | 543 | 259 | 558 | Putative alpha-1,3-mannosyltransferase MNN15 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN15 PE=2 SV=2 |
|
| 1.20e-11 | 250 | 531 | 276 | 521 | Putative alpha-1,3-mannosyltransferase MNN14 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN14 PE=2 SV=1 |
|
| 3.61e-10 | 250 | 531 | 358 | 624 | Putative alpha-1,3-mannosyltransferase MNN1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN1 PE=2 SV=1 |
|
| 2.39e-06 | 250 | 529 | 400 | 664 | Putative alpha-1,3-mannosyltransferase MNN12 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN12 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.997755 | 0.002262 |